5GPC
 
 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*AP*TP*GP*AP*GP*TP*AP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), DNA (5'-D(P*GP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*AP*CP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Transcriptional regulator (TetR/AcrR family) | | Authors: | Lee, J.Y, Yeo, H.K, Park, T.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
5GP9
 
 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | GLYCEROL, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Lee, J.Y, Yeo, H.K, Park, Y.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
3PD7
 
 | |
3QJM
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
5FUU
 
 | | Ectodomain of cleaved wild type JR-FL EnvdCT trimer in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Cryo-Em Structure of a Native, Fully Glycosylated and Cleaved HIV-1 Envelope Trimer
Science, 351, 2016
|
|
4O6J
 
 | | Crystal sturucture of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5V
 
 | | Crystal structure of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LQ8
 
 | |
3QJN
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
2G3R
 
 | | Crystal Structure of 53BP1 tandem tudor domains at 1.2 A resolution | | Descriptor: | SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
5XWB
 
 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
3VF0
 
 | | Raver1 in complex with metavinculin L954 deletion mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ribonucleoprotein PTB-binding 1, ... | | Authors: | Lee, J.H, Vonrhein, C, Bricogne, G, Izard, T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Metavinculin Tail Domain Directs Constitutive Interactions with Raver1 and vinculin RNA.
J.Mol.Biol., 422, 2012
|
|
3UYU
 
 | | Structural basis for the antifreeze activity of an ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, GLYCEROL | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
7WBK
 
 | |
7WBM
 
 | | Crystal structure of Legionella pneumophila effector protein Lpg0081 | | Descriptor: | Lpg0081, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Lee, J, Kim, H, Oh, B.H. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible modification of mitochondrial ADP/ATP translocases by paired Legionella effector proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3O49
 
 | |
3OL0
 
 | |
3Q8W
 
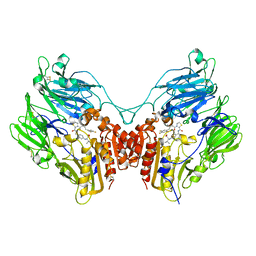 | | A b-aminoacyl containing thiazolidine derivative and DPPIV complex | | Descriptor: | Dipeptidyl peptidase 4, N-(4-{[({(2R)-3-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]-1,3-thiazolidin-2-yl}carbonyl)amino]methyl}phenyl)-D-valine | | Authors: | Lee, J.O, Song, D.H. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Discovery of b-aminoacyl containing thiazolidine derivatives as potent and selective dipeptidyl peptidase IV inhibitors
To be Published
|
|
6KHD
 
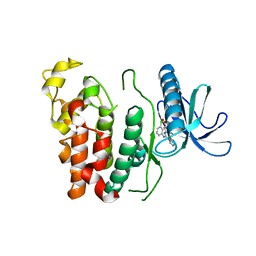 | | Crystal structure of CLK1 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK1 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
6KHE
 
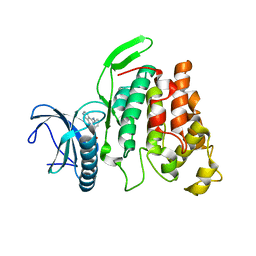 | | Crystal structure of CLK2 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
6KHF
 
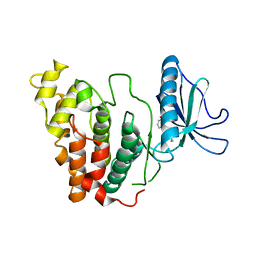 | | Crystal structure of CLK3 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
3UYV
 
 | | Crystal structure of a glycosylated ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
2F2B
 
 | | Crystal structure of integral membrane protein Aquaporin AqpM at 1.68A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3OGF
 
 | |
2LXJ
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp with dT7 | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
