5BVT
 
 | | Palmitate-bound pFABP5 | | Descriptor: | Epidermal fatty acid-binding protein, PALMITOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
7YLK
 
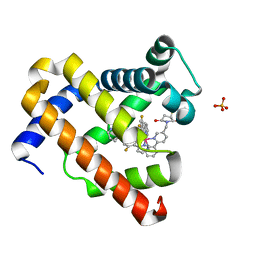 | | Myoglobin containing Ir complex | | Descriptor: | Myoglobin, SULFATE ION, delta-{1-([2,2'-bipyridin]-5-ylmethyl)pyrrolidine-2,5-dione}bis[2-(2,4-difluorophenyl)pyridine)]iridium(III), ... | | Authors: | Lee, J.H, Song, W.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Photocatalytic C-O Coupling Enzymes That Operate via Intramolecular Electron Transfer.
J.Am.Chem.Soc., 145, 2023
|
|
1X3Z
 
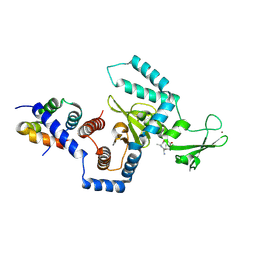 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8K07
 
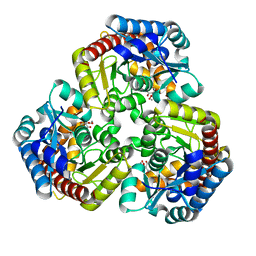 | |
1X3W
 
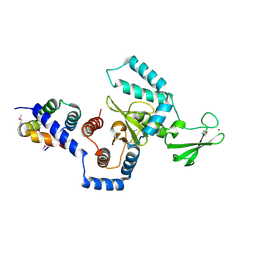 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8K06
 
 | | Pseudouridine 5'-monophosphate glycosylase from Arabidopsis thaliana -- PSU, R5P bound K185A mutant | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lee, J.Y, Kim, S.H, Rhee, S.K. | | Deposit date: | 2023-07-07 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structure and function of the pseudouridine 5'-monophosphate glycosylase PUMY from Arabidopsis thaliana.
Rna Biol., 21, 2024
|
|
8HMW
 
 | |
6JQS
 
 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
8K05
 
 | |
2FB9
 
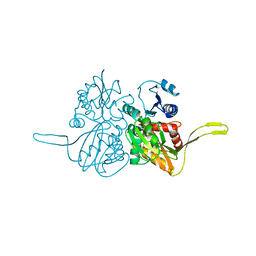 | |
6IFH
 
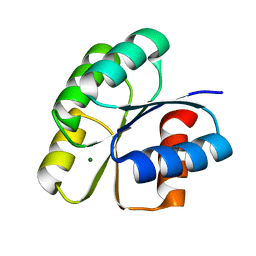 | | Unphosphorylated Spo0F from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase F | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unphosphorylated Spo0F from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
Biodesign, 6(4), 2019
|
|
7WB3
 
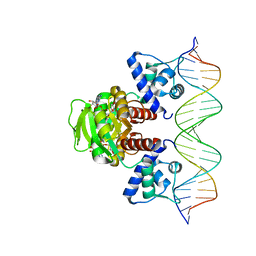 | | Crystal structure of T. maritima Rex in ternary complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*GP*AP*GP*AP*AP*AP*TP*TP*TP*AP*TP*CP*AP*CP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*TP*GP*AP*TP*AP*AP*AP*TP*TP*TP*CP*TP*CP*AP*AP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lee, J.Y, Jeong, K.H, Lee, H.J, Park, Y.W. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis of Redox-Sensing Transcriptional Repressor Rex with Cofactor NAD + and Operator DNA.
Int J Mol Sci, 23, 2022
|
|
5XGX
 
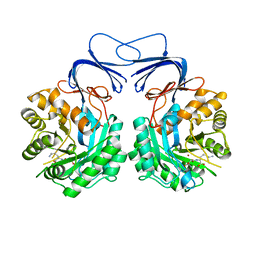 | | Crystal structure of colwellia psychrerythraea strain 34H isoaspartyl dipeptidase E80Q mutant complexed with beta-isoaspartyl lysine | | Descriptor: | D-ASPARTIC ACID, D-LYSINE, Isoaspartyl dipeptidase, ... | | Authors: | Lee, J.H, Lee, C.W, Park, S.H. | | Deposit date: | 2017-04-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and functional characterization of an isoaspartyl dipeptidase (CpsIadA) from Colwellia psychrerythraea strain 34H.
PLoS ONE, 12, 2017
|
|
3QD6
 
 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
1DGS
 
 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
5XGW
 
 | |
5BY2
 
 | |
1IDO
 
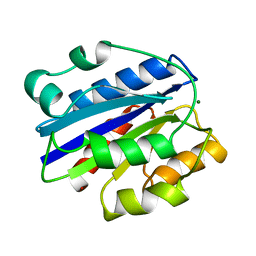 | | I-DOMAIN FROM INTEGRIN CR3, MG2+ BOUND | | Descriptor: | INTEGRIN, MAGNESIUM ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-03-12 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18).
Cell(Cambridge,Mass.), 80, 1995
|
|
6K65
 
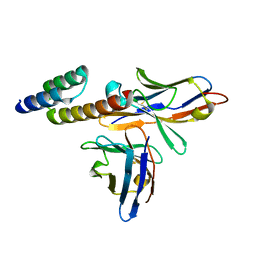 | | Application of anti-helix antibodies in protein structure determination (9014-1P4B) | | Descriptor: | 1P4B variable heavy chain, 1P4B variable light chain, Immunoglobulin G-binding protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6B
 
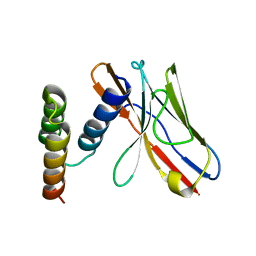 | | Application of anti-helix antibodies in protein structure determination (8496-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K3M
 
 | | Application of anti-helix antibodies in protein structure determination (8189-3LRH) | | Descriptor: | 3LRH intrabody, SpA IgG-binding domain protein,Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K67
 
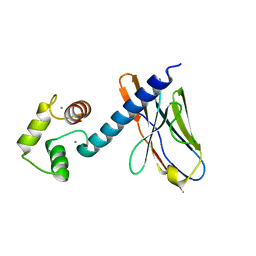 | | Application of anti-helix antibodies in protein structure determination (9011-3LRH) | | Descriptor: | 3LRH introbody, CALCIUM ION, Engineered calmodulin | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5Z2E
 
 | |
1JLM
 
 | | I-DOMAIN FROM INTEGRIN CR3, MN2+ BOUND | | Descriptor: | INTEGRIN, MANGANESE (II) ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-04-09 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two conformations of the integrin A-domain (I-domain): a pathway for activation?
Structure, 3, 1995
|
|
5Z2F
 
 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
