5D50
 
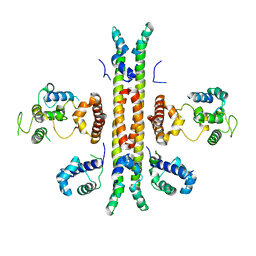 | | Crystal structure of Rep-Ant complex from Salmonella-temperate phage | | Descriptor: | Anti-repressor protein, Repressor | | Authors: | Son, S.H, Yoon, H.J, Ryu, S, Lee, H.H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUU
 
 | |
8GVW
 
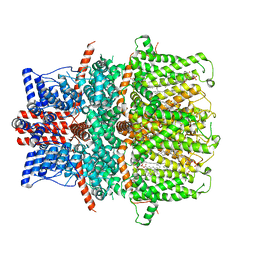 | | Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class2 | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
8GVX
 
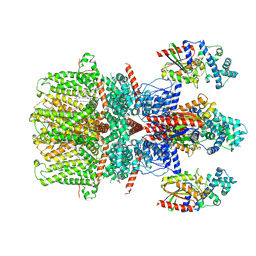 | | Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class2 | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Molecular architecture of the G alpha i -bound TRPC5 ion channel.
Nat Commun, 14, 2023
|
|
7VOV
 
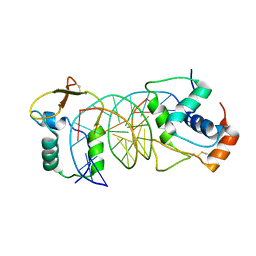 | | The crystal structure of human forkhead box protein in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOX
 
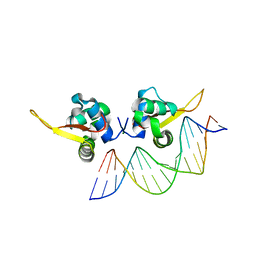 | | The crystal structure of human forkhead box protein A in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*TP*T)-3'), Hepatocyte nuclear factor 3-alpha, ... | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOU
 
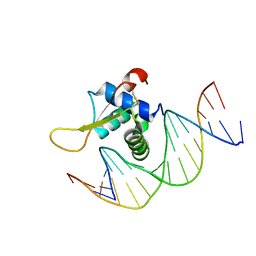 | | The crystal structure of human forkhead box protein in complex with DNA 1 | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
8HST
 
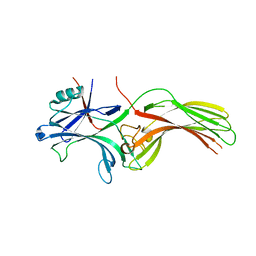 | |
8HSV
 
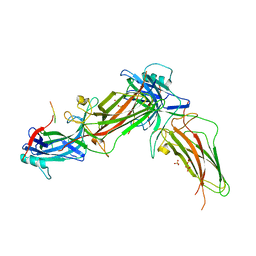 | | The structure of rat beta-arrestin1 in complex with a rat Mdm2 peptide | | Descriptor: | Beta-arrestin-1, SULFATE ION, peptide from E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yun, Y, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GPCR targeting of E3 ubiquitin ligase MDM2 by inactive beta-arrestin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2O4C
 
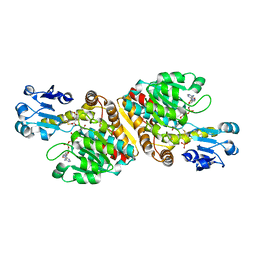 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
3W8L
 
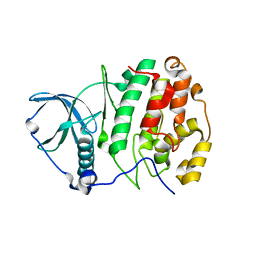 | | Crystal structure of human CK2 in complex with inositol hexakisphosphate | | Descriptor: | Casein kinase II subunit alpha, INOSITOL HEXAKISPHOSPHATE | | Authors: | Son, S.H, Lee, W.-K, Yu, Y.G, Lee, H.H. | | Deposit date: | 2013-03-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8WPL
 
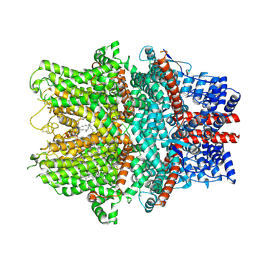 | |
8WPN
 
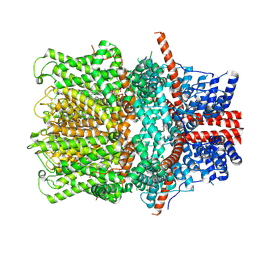 | |
8WPM
 
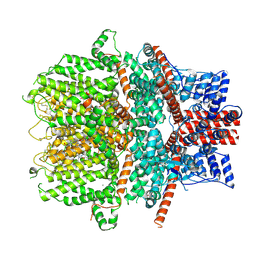 | |
7E64
 
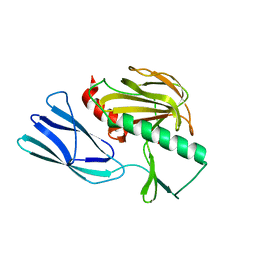 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
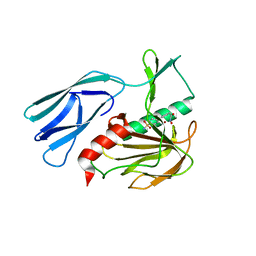 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | Descriptor: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
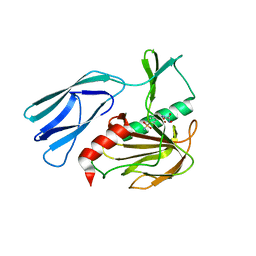 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | Descriptor: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
6JN8
 
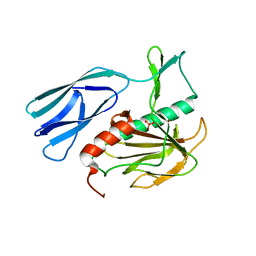 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
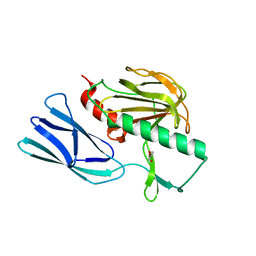 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
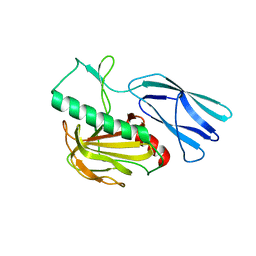 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
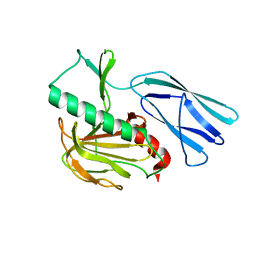 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
