8T6B
 
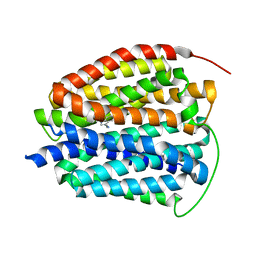 | | Human VMAT2 in complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T69
 
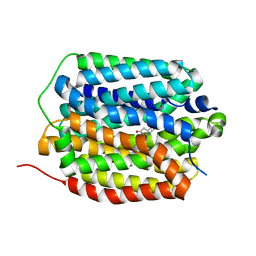 | | Human VMAT2 in complex with tetrabenazine | | Descriptor: | Synaptic vesicular amine transporter, tetrabenazine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T6A
 
 | | Human VMAT2 in complex with reserpine | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
1X3Z
 
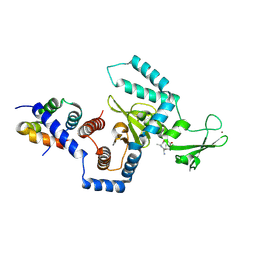 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6IYM
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
7XQG
 
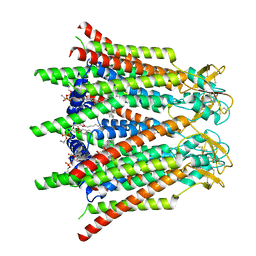 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN conformation) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQH
 
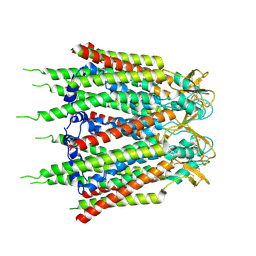 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN-TM1i conformation) | | Descriptor: | C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XRI
 
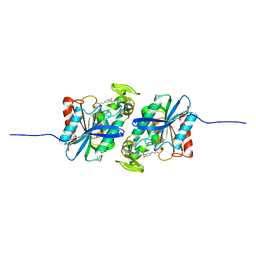 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
2DF7
 
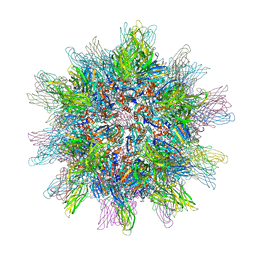 | | Crystal structure of infectious bursal disease virus VP2 subviral particle | | Descriptor: | CALCIUM ION, CHLORIDE ION, structural polyprotein VP2 | | Authors: | Ko, T.P, Lee, C.C, Wang, M.Y, Wang, A.H. | | Deposit date: | 2006-02-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6A resolution: Implications in virion assembly and immunogenicity.
J.Struct.Biol., 155, 2006
|
|
1X3W
 
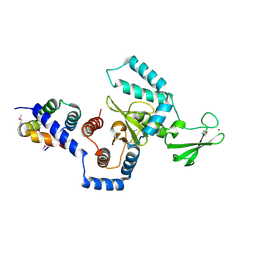 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5XJG
 
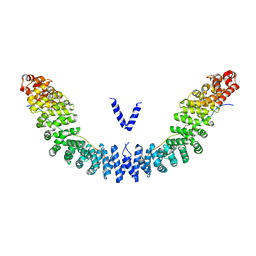 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8D36
 
 | |
7UKC
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 1, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKH
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, intracellular region | | Descriptor: | CALCIUM ION, Isoform 2 of Kv channel-interacting protein 2, Potassium voltage-gated channel subfamily D member 2, ... | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKE
 
 | |
7UKG
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, transmembrane region | | Descriptor: | Dipeptidyl-peptidase 6, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UK5
 
 | |
7UKD
 
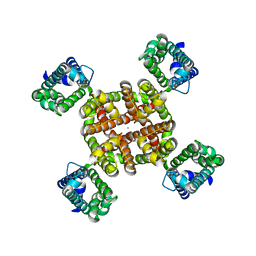 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 2, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKF
 
 | | Human Kv4.2-KChIP2 channel complex in a putative resting state, transmembrane region | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7XQJ
 
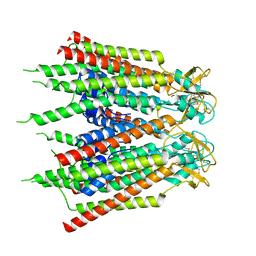 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQI
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQF
 
 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
