1ATG
 
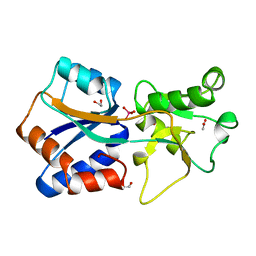 | | AZOTOBACTER VINELANDII PERIPLASMIC MOLYBDATE-BINDING PROTEIN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PERIPLASMIC MOLYBDATE-BINDING PROTEIN, ... | | Authors: | Lawson, D.M, Pau, R.N, Williams, C.E.M, Mitchenall, L.A. | | Deposit date: | 1997-08-14 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA.
Structure, 6, 1998
|
|
1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1E84
 
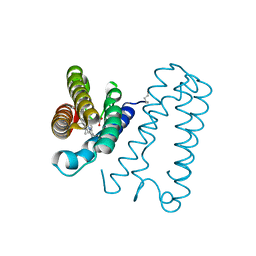 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E83
 
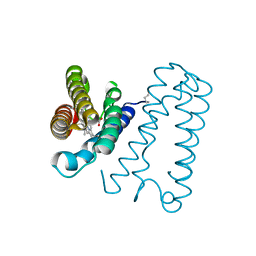 | | Cytochrome c' from Alcaligenes xylosoxidans - oxidized structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E86
 
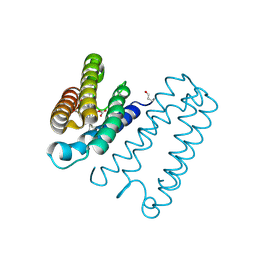 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with CO bound to distal side of heme | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E85
 
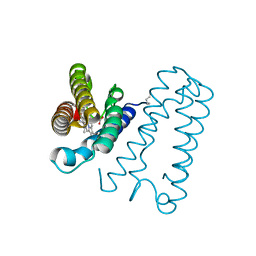 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with NO bound to proximal side of heme | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase.
Embo J., 19, 2000
|
|
6BL8
 
 | |
4XAQ
 
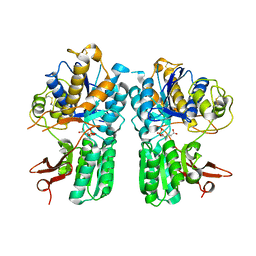 | | mGluR2 ECD and mGluR3 ECD with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, CHLORIDE ION, Metabotropic glutamate receptor 2, ... | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
4XAR
 
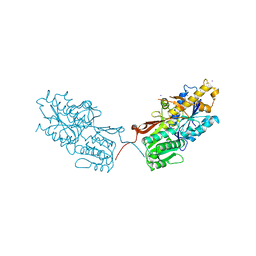 | | mGluR2 ECD and mGluR3 ECD complex with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, IODIDE ION, Metabotropic glutamate receptor 3 | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
4XAS
 
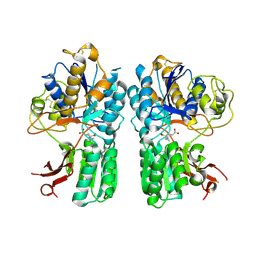 | | mGluR2 ECD ligand complex | | Descriptor: | (1R,4S,5S,6S)-4-aminospiro[bicyclo[3.1.0]hexane-2,1'-cyclopropane]-4,6-dicarboxylic acid, Metabotropic glutamate receptor 2 | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
5CNI
 
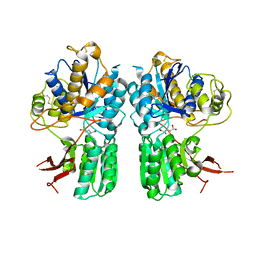 | | mGlu2 with Glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Clawson, D.K, Atwell, S, Monn, J.A. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
4TGL
 
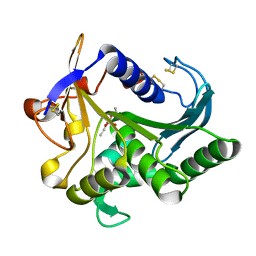 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
9FOY
 
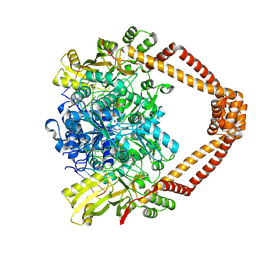 | | Ternary complex of a Mycobacterium tuberculosis DNA gyrase core fusion with DNA and the inhibitor AMK32b | | Descriptor: | (1~{R})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, (1~{S})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), ... | | Authors: | Kokot, M, Hrast, M, Feng, L, Mitchenall, L.A, Lawson, D.M, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2024-06-12 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the Mycobacterium tuberculosis DNA gyrase-DNA-NBTI complex
To be published
|
|
6TVP
 
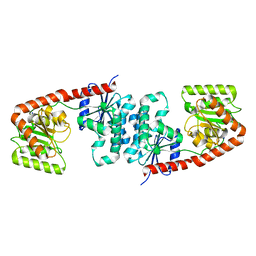 | | Structure of Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM | | Descriptor: | Alpha-maltose-1-phosphate synthase, SODIUM ION | | Authors: | Syson, K, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1QGU
 
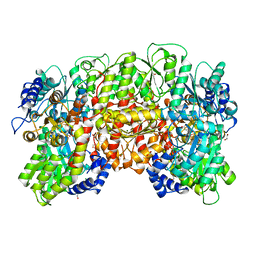 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, DITHIONITE-REDUCED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-06 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
3ZMD
 
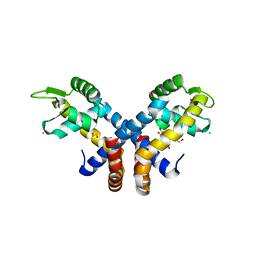 | | Crystal structure of AbsC, a MarR family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Stevenson, C.E.M, Kock, H, Mootien, S, Davies, S.C, Bibb, M.J, Lawson, D.M. | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Absc, a Marr Family Transcriptional Regulator from Streptomyces Coelicolor
To be Published
|
|
1H4D
 
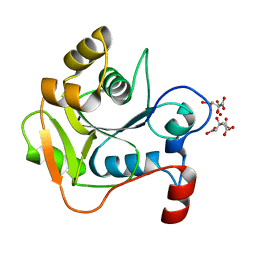 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1H4E
 
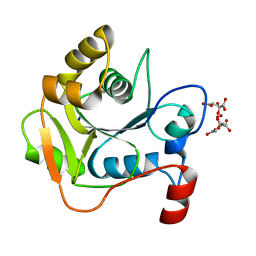 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1H4C
 
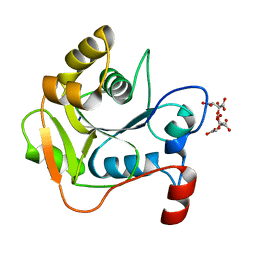 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1H1L
 
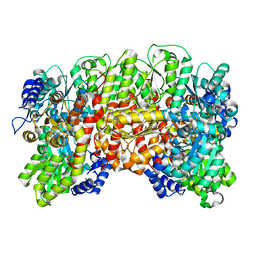 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, NIFV MUTANT | | Descriptor: | CHLORIDE ION, CITRIC ACID, FE(8)-S(7) CLUSTER, ... | | Authors: | Mayer, S.M, Gormal, C.A, Smith, B.E, Lawson, D.M. | | Deposit date: | 2002-07-18 | | Release date: | 2002-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis of the Mofe Protein of Nitrogenase from a Nifv Mutant of Klebsiella Pneumoniae Identifies Citrate as a Ligand to the Molybdenum of Iron Molybdenum Cofactor (Femoco).
J.Biol.Chem., 277, 2002
|
|
3ZSS
 
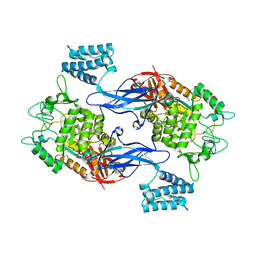 | | Apo form of GlgE isoform 1 from Streptomyces coelicolor | | Descriptor: | PUTATIVE GLUCANOHYDROLASE PEP1A | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZQL
 
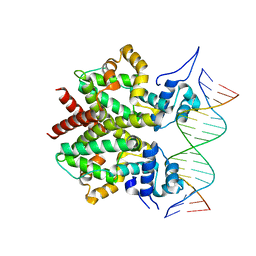 | | DNA-bound form of TetR-like repressor SimR | | Descriptor: | 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*CP*CP*GP*TP*AP*DCP *GP*AP*A)-3', 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*GP*CP*GP*TP*AP*DCP *GP*AP*A)-3', PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Schumacher, M.A, Lawson, D.M, Brennan, R.G, Buttner, M.J. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Crystal Structure of the Tetr Family Transcriptional Repressor Simr Bound to DNA and the Role of a Flexible N-Terminal Extension in Minor Groove Binding.
Nucleic Acids Res., 39, 2011
|
|
3ZST
 
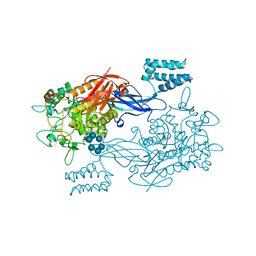 | | GlgE isoform 1 from Streptomyces coelicolor with alpha-cyclodextrin bound | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A GLGE ISOFORM 1 | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
1H9M
 
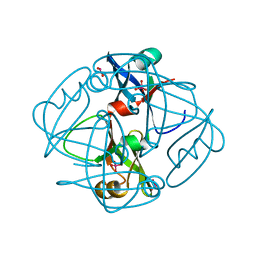 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. PEG-grown form with molybdate bound | | Descriptor: | MOLYBDATE ION, MOLYBDENUM-BINDING-PROTEIN | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
3ZPL
 
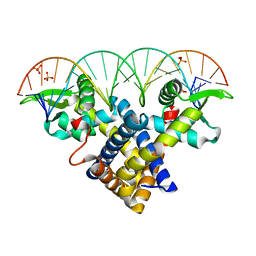 | | Crystal structure of Sco3205, a MarR family transcriptional regulator from Streptomyces coelicolor, in complex with DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*TP*GP*AP*GP*AP*TP*CP*TP *CP*AP*AP*TP*CP*TP*TP*DT)-3', PHOSPHATE ION, PUTATIVE MARR-FAMILY TRANSCRIPTIONAL REPRESSOR | | Authors: | Stevenson, C.E.M, Assaad, A, Lawson, D.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of DNA Sequence Recognition by a Streptomycete Marr Family Transcriptional Regulator Through Surface Plasmon Resonance and X-Ray Crystallography.
Nucleic Acids Res., 41, 2013
|
|
