1AFT
 
 | | SMALL SUBUNIT C-TERMINAL INHIBITORY PEPTIDE OF MOUSE RIBONUCLEOTIDE REDUCTASE AS BOUND TO THE LARGE SUBUNIT, NMR, 26 STRUCTURES | | 分子名称: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE | | 著者 | Laub, P.B, Fisher, A.L, Furst, G.T, Barwis, B.A, Hamann, C.S, Cooperman, B.S. | | 登録日 | 1997-03-13 | | 公開日 | 1997-05-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of an inhibitory R2 C-terminal peptide bound to mouse ribonucleotide reductase R1 subunit.
Nat.Struct.Biol., 2, 1995
|
|
5MM2
 
 | | nora virus structure | | 分子名称: | Capsid protein VP4A, capsid protein VP4B, capsid protein VP4C | | 著者 | Laurinmaki, P, Shakeel, S, Ekstrom, J.-O, Butcher, S.J. | | 登録日 | 2016-12-08 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structure of Nora virus at 2.7 angstrom resolution and implications for receptor binding, capsid stability and taxonomy.
Sci Rep, 10, 2020
|
|
1EQY
 
 | | COMPLEX BETWEEN RABBIT MUSCLE ALPHA-ACTIN: HUMAN GELSOLIN DOMAIN 1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ALPHA ACTIN, CALCIUM ION, ... | | 著者 | McLaughlin, P.J, Gooch, J.T, Mannherz, H.G, Weeds, A.G. | | 登録日 | 2000-04-06 | | 公開日 | 2000-05-03 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of gelsolin segment 1-actin complex and the mechanism of filament severing.
Nature, 364, 1993
|
|
4UWM
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | 分子名称: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | 登録日 | 2014-08-12 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5XKT
 
 | | Klebsiella pneumoniae UreG in complex with GMPPNP and nickel | | 分子名称: | NICKEL (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Urease accessory protein UreG | | 著者 | Fong, Y.H, Yuen, M.H, Nim, Y.S, Lau, P.H, Wong, K.B. | | 登録日 | 2017-05-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into how GTP-dependent conformational changes in a metallochaperone UreG facilitate urease maturation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3NAD
 
 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | 分子名称: | Ferulate decarboxylase, SULFATE ION | | 著者 | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | 登録日 | 2010-06-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7ZAK
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | 登録日 | 2022-03-22 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | 著者 | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
5AEC
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | 分子名称: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | 登録日 | 2015-08-28 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3UOY
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 1) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO, ... | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOV
 
 | | Crystal Structure of OTEMO (FAD bound form 1) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.045 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP5
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.453 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP4
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 3) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOZ
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 2) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.407 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOX
 
 | | Crystal Structure of OTEMO (FAD bound form 2) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.956 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
4GAG
 
 | | Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 412-423) | | 分子名称: | GLYCEROL, Genome polyprotein, NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, ... | | 著者 | Potter, J.A, Owsianka, A, Angus, A.G.N, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | 登録日 | 2012-07-25 | | 公開日 | 2012-10-10 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
4GAY
 
 | | Structure of the broadly neutralizing antibody AP33 | | 分子名称: | NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, NEUTRALIZING ANTIBODY AP33 LIGHT CHAIN, TRIETHYLENE GLYCOL | | 著者 | Potter, J.A, Owsianka, A, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | 登録日 | 2012-07-26 | | 公開日 | 2012-10-10 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | 分子名称: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | 著者 | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | 登録日 | 2000-05-24 | | 公開日 | 2000-06-21 | | 最終更新日 | 2011-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
2K77
 
 | | NMR solution structure of the Bacillus subtilis ClpC N-domain | | 分子名称: | Negative regulator of genetic competence clpC/mecB | | 著者 | Kojetin, D.J, McLaughlin, P.D, Thompson, R.J, Rance, M, Cavanagh, J. | | 登録日 | 2008-08-04 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions.
J.Mol.Biol., 387, 2009
|
|
7LOO
 
 | | S-adenosyl methionine transferase cocrystallized with ATP | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Jackson, C.J, Tan, L.L, Laurino, P. | | 登録日 | 2021-02-10 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
8X2W
 
 | |
8X2V
 
 | |
6WFS
 
 | | Cryo-EM Structure of Hepatitis B virus T=4 capsid in complex with the antiviral molecule DBT1 | | 分子名称: | 11-oxo-N-[2-(4-sulfamoylphenyl)ethyl]-10,11-dihydrodibenzo[b,f][1,4]thiazepine-8-carboxamide, Capsid protein | | 著者 | Schlicksup, C, Laughlin, P, Dunkelbarger, S, Wang, J.C, Zlotnick, A. | | 登録日 | 2020-04-03 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids.
Acs Chem.Biol., 15, 2020
|
|
8WCH
 
 | |
3J2J
 
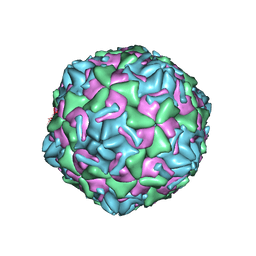 | | Empty coxsackievirus A9 capsid | | 分子名称: | Protein VP1, Protein VP2, Protein VP3 | | 著者 | Shakeel, S, Seitsonen, J.J.T, Kajander, T, Laurinmaki, P, Hyypia, T, Susi, P, Butcher, S.J. | | 登録日 | 2012-10-04 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.54 Å) | | 主引用文献 | Structural and functional analysis of coxsackievirus A9 integrin {alpha}v{beta}6 binding and uncoating.
J.Virol., 87, 2013
|
|
