6SMF
 
 | |
6SME
 
 | |
6HS9
 
 | |
6HSU
 
 | |
6HSQ
 
 | |
6HSA
 
 | |
6HS8
 
 | |
6HSR
 
 | |
6HSB
 
 | |
1HRP
 
 | | CRYSTAL STRUCTURE OF HUMAN CHORIONIC GONADOTROPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN CHORIONIC GONADOTROPIN | | Authors: | Lapthorn, A.J, Harris, D.C, Isaacs, N.W. | | Deposit date: | 1994-08-15 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human chorionic gonadotropin.
Nature, 369, 1994
|
|
1SHK
 
 | |
2BT4
 
 | | Type II Dehydroquinase inhibitor complex | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Toscano, M.D, Stewart, K.A, Coggins, J.R, Lapthorn, A.J, Abell, C. | | Deposit date: | 2005-05-26 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design of New Bifunctional Inhibitors of Type II Dehydroquinase.
Org.Biomol.Chem., 3, 2005
|
|
1D0I
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR COMPLEXED WITH PHOSPHATE IONS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, TYPE II 3-DEHYDROQUINATE HYDRATASE | | Authors: | Roszak, A.W, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1999-09-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor.
Structure, 10, 2002
|
|
2C4W
 
 | |
2C57
 
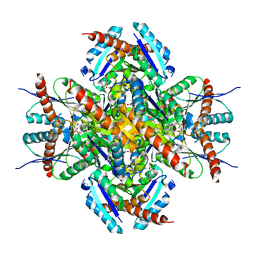 | | H.pylori type II dehydroquinase in complex with FA1 | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Robinson, D.A, Lapthorn, A.J. | | Deposit date: | 2005-10-26 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Helicobacter Pylori Type II Dehydroquinase Inhibitor Complexes: New Directions for Inhibitor Design.
J.Med.Chem., 49, 2006
|
|
2C4V
 
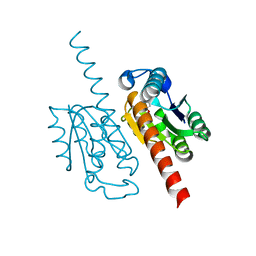 | | H. pylori type II DHQase in complex with citrate | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, CITRIC ACID | | Authors: | Robinson, D.A, Lapthorn, A.J. | | Deposit date: | 2005-10-24 | | Release date: | 2006-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Helicobacter Pylori Type II Dehydroquinase Inhibitor Complexes: New Directions for Inhibitor Design.
J.Med.Chem., 49, 2006
|
|
1V1J
 
 | | Crystal structure of type II Dehydroquintae Dehydratase from Streptomyces coelicolor in complex with 3-fluoro | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ANHYDRO-3-FLUORO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2004-04-16 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | (1R,4S,5R)-3-Fluoro-1,4,5-Trihydroxy-2-Cyclohexene-1-Carboxylic Acid: The Fluoro Analogue of the Enolate Intermediate in the Reaction Catalyzed by Type II Dehydroquinases
Org.Biomol.Chem., 2, 2004
|
|
4AST
 
 | |
1BGS
 
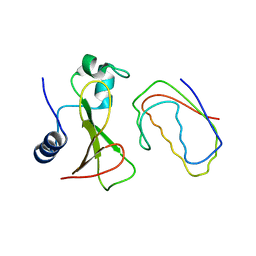 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
4AUB
 
 | |
5EL8
 
 | |
5ELA
 
 | |
2VD3
 
 | | The structure of histidine inhibited HisG from Methanobacterium thermoautotrophicum | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ATP PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Lohkamp, B, Schweikert, T, Lapthorn, A.J. | | Deposit date: | 2007-09-28 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of Histidine Inhibited Hisg from Methanobacterium Thermoautotrophicum
To be Published
|
|
2CJF
 
 | | TYPE II DEHYDROQUINASE INHIBITOR COMPLEX | | Descriptor: | (1S,4S,5S)-1,4,5-TRIHYDROXY-3-[3-(PHENYLTHIO)PHENYL]CYCLOHEX-2-ENE-1-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Payne, R.J, Riboldi-Tunnicliffe, A, Abell, A.D, Lapthorn, A.J, Abell, C. | | Deposit date: | 2006-03-31 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, and structural studies on potent biaryl inhibitors of type II dehydroquinases.
Chemmedchem, 2, 2007
|
|
5ELG
 
 | |
