8CWI
 
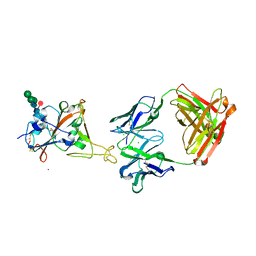 | | Fab arm of antibody 10G4 bound to CoV-2 receptor binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of Fab arm of antibody 10G4, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Neutralization of CoV-2 omicron lineages by affinity-matured class 5 antibodies
To Be Published
|
|
8CWJ
 
 | |
8CWK
 
 | |
8DXU
 
 | |
8DXT
 
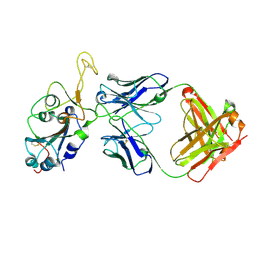 | | Fab arm of antibody GAR12 bound to the receptor binding domain of SARS-CoV-2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab arm of antibody GAR12, Light chain of Fab arm of antibody GAR12, ... | | Authors: | Langley, D.B, Christ, D, Henry, J.Y. | | Deposit date: | 2022-08-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
7MSQ
 
 | |
7UEL
 
 | |
7UEM
 
 | |
7UEN
 
 | |
4X4X
 
 | |
4X4Z
 
 | |
7KZB
 
 | |
7KZC
 
 | |
7KZA
 
 | |
2ZCG
 
 | |
2ZP2
 
 | |
1DUV
 
 | | CRYSTAL STRUCTURE OF E. COLI ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH NDELTA-L-ORNITHINE-DIAMINOPHOSPHINYL-N-SULPHONIC ACID (PSORN) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDELTA-(N'-SULPHODIAMINOPHOSPHINYL)-L-ORNITHINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Langley, D.B, Templeton, M.D, Fields, B.A, Mitchell, R.E, Collyer, C.A. | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of ornithine transcarbamoylase by Ndelta -(N'-Sulfodiaminophosphinyl)-L-ornithine, a true transition state analogue? Crystal structure and implications for catalytic mechanism.
J.Biol.Chem., 275, 2000
|
|
7RTA
 
 | | Crystal structures of human PYY and NPY | | Descriptor: | 4A3B2-B Fab heavy chain, 4A3B2-B Fab light chain, Neuropeptide Y, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2021-08-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human neuropeptide Y (NPY) and peptide YY (PYY).
Neuropeptides, 92, 2022
|
|
7RT9
 
 | | Crystal structures of human PYY and NPY | | Descriptor: | 4A3B2-A Fab heavy chain, 4A3B2-A Fab light chain, GLYCEROL, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2021-08-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human neuropeptide Y (NPY) and peptide YY (PYY).
Neuropeptides, 92, 2022
|
|
7T72
 
 | | Epitope-based selection of SARS-CoV-2 neutralizing antibodies from convalescent patients | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody heavy chain, Antibody light chain, ... | | Authors: | Langley, D.B, Christ, D, Rouet, R. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.177 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
6P4B
 
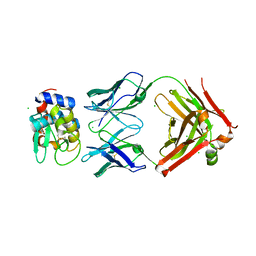 | | HyHEL10 fab variant HyHEL10-4x (heavy chain mutations L4F, Y33H, S56N, and Y58F) bound to hen egg lysozyme variant HEL2x-flex (mutations R21Q, R73E, C76S, and C94S) | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4C
 
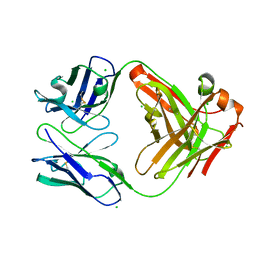 | | HyHEL10 Fab carrying four heavy chain mutations (HyHEL10-4x): L4F, Y33H, S56N, and Y58F | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4D
 
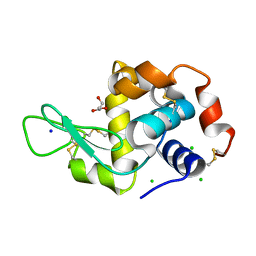 | | Hen egg lysozyme (HEL) containing three point mutations (HEL3x): R21Q, R73E, and D101R | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4A
 
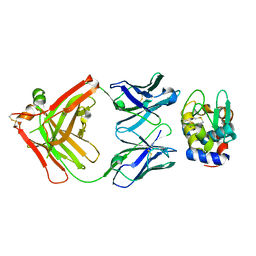 | |
2CFD
 
 | | AGAO in complex with wc4l3 (Ru-wire inhibitor, 4-carbon linker, lambda enantiomer, data set 3) | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-20 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
