6P4P
 
 | | Salmonella typhi PltB Homopentamer N29K Mutant | | Descriptor: | Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
5JH8
 
 | | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472 | | Descriptor: | (2S)-2-(dimethylamino)-4-(methylselanyl)butanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chang, C, Michalska, K, Tesar, C, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.018 Å) | | Cite: | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472
To Be Published
|
|
4H7N
 
 | |
3MVN
 
 | | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP | | Descriptor: | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP
To be Published
|
|
3K2N
 
 | |
5CCS
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
5CJ3
 
 | | Crystal structure of the zorbamycin binding protein (ZbmA) from Streptomyces flavoviridis with zorbamycin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Zbm binding protein, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Rudolf, J.D, Ma, M, Chang, C.-Y, Lohman, J.R, Yang, D, Shen, B, Enzyme Discovery for Natural Product Biosynthesis, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-13 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
8B81
 
 | | The structure of Gan1D W433A in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | The structure of Gan1D W433A in complex with cellobiose-6-phosphate
To Be Published
|
|
8B80
 
 | | The structure of Gan1D W433A in complex with galactose-6P | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of Gan1D W433A in complex with galactose-6P
To Be Published
|
|
4RA7
 
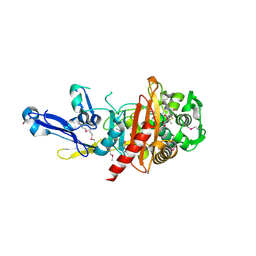 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-2-hydroxy-1-{[(2-hydroxynaphthalen-1-yl)carbonyl]amino}ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin
To be Published
|
|
4MQD
 
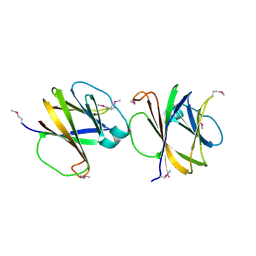 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | Descriptor: | DNA-entry nuclease inhibitor | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
4E9C
 
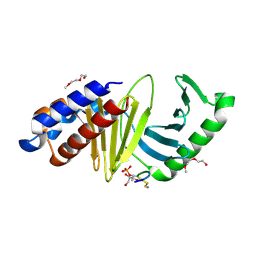 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with LDPPLHSpTA phosphopeptide | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, LDPPLHSpTA phosphopeptide, ... | | Authors: | Sledz, P, Hyvonen, M, Lang, S, Stubbs, C.J, Abell, C. | | Deposit date: | 2012-03-21 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-throughput interrogation of ligand binding mode using a fluorescence-based assay.
Angew. Chem. Int. Ed. Engl., 51, 2012
|
|
4GS5
 
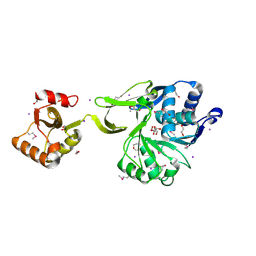 | | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein, IODIDE ION | | Authors: | Tan, K, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053
To be Published
|
|
6GT9
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose | | Descriptor: | Putative sugar binding protein, SULFATE ION, beta-D-galactopyranose | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-16 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose
To Be Published
|
|
4RM1
 
 | |
6XUI
 
 | | Crystal structure of human phosphoglucose isomerase in complex with inhibitor | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5-PHOSPHOARABINONIC ACID, GLYCEROL, ... | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2020-01-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Bioorg.Chem., 102, 2020
|
|
3KDQ
 
 | | Crystal structure of a functionally unknown conserved protein from Corynebacterium diphtheriae. | | Descriptor: | uncharacterized conserved protein | | Authors: | Zhang, R, Wu, R, Tan, K, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a functionally unknown conserved protein from Corynebacterium diphtheriae.
To be Published
|
|
5AEQ
 
 | | Neuronal calcium sensor (NCS-1)from Rattus norvegicus | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, SODIUM ION | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5JMU
 
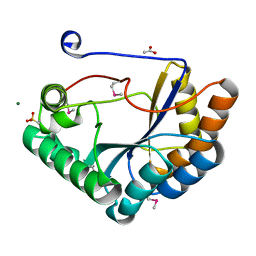 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
5CBV
 
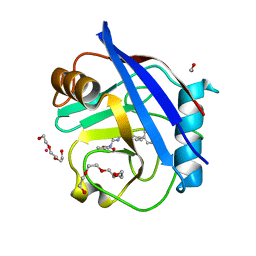 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, Human Cyclophilin D, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
3B4Q
 
 | |
5U0S
 
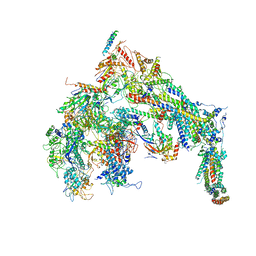 | | Cryo-EM structure of the Mediator-RNAPII complex | | Descriptor: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | Authors: | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | Deposit date: | 2016-11-26 | | Release date: | 2017-03-08 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
5CBW
 
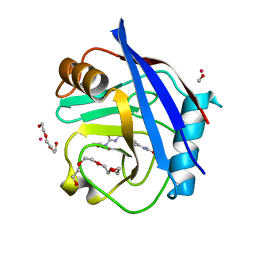 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
3BJN
 
 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
4R0Q
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cephalothin | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CEPHALOTHIN GROUP, Peptidoglycan glycosyltransferase, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-01 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cephalothin
To be Published
|
|
