2HEI
 
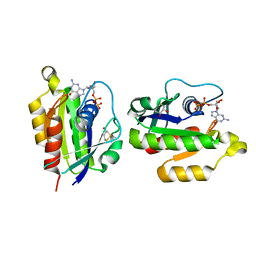 | | Crystal structure of human RAB5B in complex with GDP | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5B | | Authors: | Hong, B, Shen, L, Wang, J, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human RAB5B in complex with GDP
To be Published
|
|
2I7Q
 
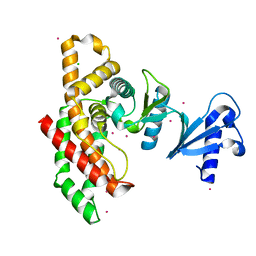 | | Crystal structure of Human Choline Kinase A | | Descriptor: | CHLORIDE ION, Choline kinase alpha, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Wasney, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Human Choline Kinase A
To be Published
|
|
2P5S
 
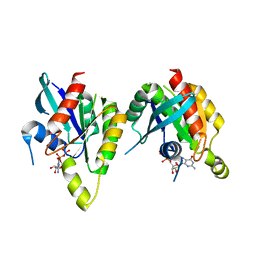 | | RAB domain of human RASEF in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, RAS and EF-hand domain containing, UNKNOWN ATOM OR ION | | Authors: | Zhu, H, Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | RAB domain of human RASEF in complex with GDP
To be Published
|
|
2KX9
 
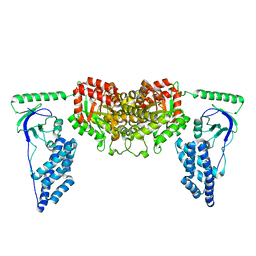 | | Solution Structure of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Schwieters, C.D, Suh, J, Grishaev, A, Takayama, Y, Guirlando, R, Clore, G. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2GRY
 
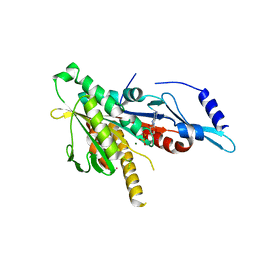 | | Crystal structure of the human KIF2 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human KIF2 motor domain in complex with ADP
to be published
|
|
2QFZ
 
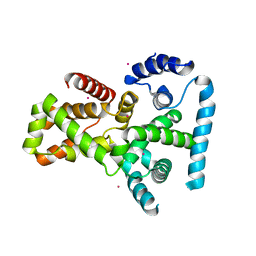 | | Crystal structure of human TBC1 domain family member 22A | | Descriptor: | TBC1 domain family member 22A, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Dimov, S, Dong, A, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human TBC1 domain family member 22A.
To be Published
|
|
2L5H
 
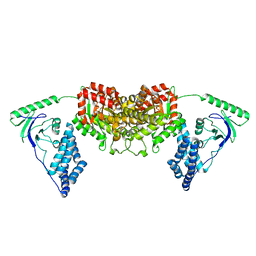 | | Solution Structure of the H189Q mutant of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Takayama, Y.D, Schwieters, C.D, Grishaev, A, Guirlando, R, Clore, G. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Combined Use of Residual Dipolar Couplings and Solution X-ray Scattering To Rapidly Probe Rigid-Body Conformational Transitions in a Non-phosphorylatable Active-Site Mutant of the 128 kDa Enzyme I Dimer.
J.Am.Chem.Soc., 133, 2011
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2Y8Q
 
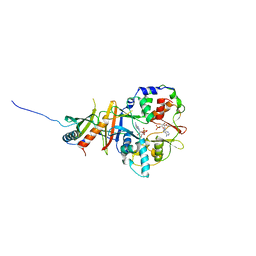 | | Structure of the regulatory fragment of mammalian AMPK in complex with one ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2Y8L
 
 | | Structure of the regulatory fragment of mammalian aMPK in complex with two ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2YA3
 
 | | STRUCTURE OF THE REGULATORY FRAGMENT OF MAMMALIAN AMPK IN COMPLEX WITH COUMARIN ADP | | Descriptor: | 3'-(7-diethylaminocoumarin-3-carbonylamino)-3'-deoxy-ADP, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
6T26
 
 | | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Asgeirsson, B, Hjorleifsson, J.G, Markusson, S, Helland, R. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine.
Biochem Biophys Rep, 24, 2020
|
|
7BJK
 
 | |
1SAX
 
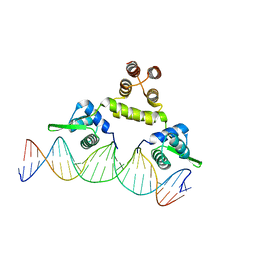 | | Three-dimensional structure of s.aureus methicillin-resistance regulating transcriptional repressor meci in complex with 25-bp ds-DNA | | Descriptor: | 5'-d(CAAAATTACAACTGTAATATCGGAG)-3', 5'-d(GCTCCGATATTACAGTTGTAATTTT)-3', Methicillin resistance regulatory protein mecI, ... | | Authors: | Garcia-Castellanos, R, Mallorqui-Fernandez, G, Marrero, A, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the transcriptional regulation of methicillin resistance: MecI repressor in complex with its operator
J.Biol.Chem., 279, 2004
|
|
2ISA
 
 | | Crystal Structure of Vibrio salmonicida catalase | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Riise, E.K, Lorentzen, M.S, Helland, R, Smalas, A.O, Leiros, H.K.S, Willassen, N.P. | | Deposit date: | 2006-10-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The first structure of a cold-active catalase from Vibrio salmonicida at 1.96A reveals structural aspects of cold adaptation
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2M8L
 
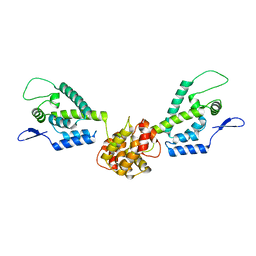 | | HIV capsid dimer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-23 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M8P
 
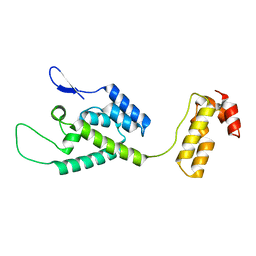 | | The structure of the W184AM185A mutant of the HIV-1 capsid protein | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2LY8
 
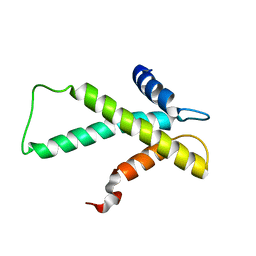 | | The budding yeast chaperone Scm3 recognizes the partially unfolded dimer of the centromere-specific Cse4/H4 histone variant | | Descriptor: | Budding yeast chaperone Scm3 | | Authors: | Hong, J, Feng, H, Zhou, Z, Ghirlando, R, Bai, Y. | | Deposit date: | 2012-09-13 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of Functionally Conserved Regions in the Structure of the Chaperone/CenH3/H4 Complex.
J.Mol.Biol., 425, 2013
|
|
2M8N
 
 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
5G3H
 
 | | Preserving Metallic Sites Affected by Radiation Damage the CuT2 Case in Thermus Thermophilus Multicopper oxidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Ruiz-Arellano, R, Diaz, A, Rosas, E, Rudino, E. | | Deposit date: | 2016-04-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Preserving Metallic Sites Affected by Radiation Damage the Cut2 Case in Thermus Thermophilus Multicopper Oxidase
To be Published
|
|
1X9Z
 
 | | Crystal structure of the MutL C-terminal domain | | Descriptor: | CHLORIDE ION, DNA mismatch repair protein mutL, GLYCEROL, ... | | Authors: | Guarne, A, Ramon-Maiques, S, Wolff, E.M, Ghirlando, R, Hu, X, Miller, J.H, Yang, W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair
Embo J., 23, 2004
|
|
5MY1
 
 | | E. coli expressome | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kohler, R, Mooney, R.A, Mills, D.J, Kostrewa, D, Landick, R, Cramer, P. | | Deposit date: | 2017-01-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Architecture of a transcribing-translating expressome.
Science, 356, 2017
|
|
5G3E
 
 | | Preserving Metallic Sites Affected by Radiation DAmage the CuT2 CAse in THermus Thermophilus Multicopper Oxidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Ruiz-Arellano, R, Diaz, A, Rosas, E, Rudino, E. | | Deposit date: | 2016-04-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Preserving Metallic Sites Affected by Radiation Damage the Cut2 Case in Thermus Thermophilus Multicopper Oxidase
To be Published
|
|
2BW3
 
 | | Three-dimensional structure of the Hermes DNA transposase | | Descriptor: | TRANSPOSASE | | Authors: | Hickman, A.B, Perez, Z.N, Zhou, L, Musingarimi, P, Ghirlando, R, Hinshaw, J.E, Craig, N.L, Dyda, F. | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of a Eukaryotic DNA Transposase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2E2D
 
 | | Flexibility and variability of TIMP binding: X-ray structure of the complex between collagenase-3/MMP-13 and TIMP-2 | | Descriptor: | CALCIUM ION, Matrix metallopeptidase 13, Metalloproteinase inhibitor 2, ... | | Authors: | Maskos, K, Lang, R, Tschesche, H, Bode, W. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility and Variability of TIMP Binding: X-ray Structure of the Complex Between Collagenase-3/MMP-13 and TIMP-2
J.Mol.Biol., 366, 2007
|
|
