3ST9
 
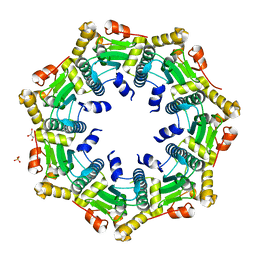 | | Crystal structure of ClpP in heptameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
4HBL
 
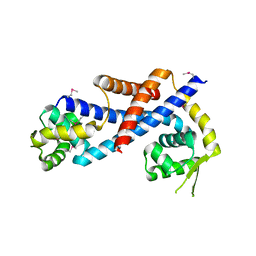 | | Crystal structure of AbfR of Staphylococcus epidermidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Liu, X, Sun, X, Gan, J, Lan, L, Yang, C.-G. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-02 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation-sensing Regulator AbfR Regulates Oxidative Stress Responses, Bacterial Aggregation, and Biofilm Formation in Staphylococcus epidermidis.
J.Biol.Chem., 288, 2013
|
|
4V9J
 
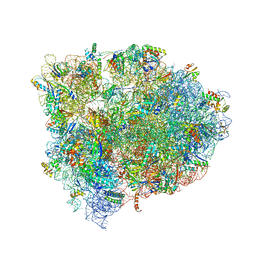 | | 70S ribosome translocation intermediate GDPNP-II containing elongation factor EFG/GDPNP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.86 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
4V89
 
 | | Crystal Structure of Release Factor RF3 Trapped in the GTP State on a Rotated Conformation of the Ribosome (without viomycin) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Trakhanov, S, Noller, H.F. | | Deposit date: | 2011-11-17 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome.
Rna, 18, 2012
|
|
4W29
 
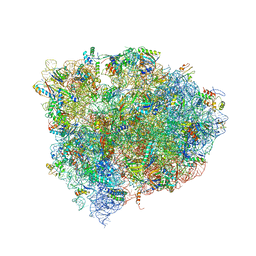 | | 70S ribosome translocation intermediate containing elongation factor EFG/GDP/fusidic acid, mRNA, and tRNAs trapped in the AP/AP pe/E chimeric hybrid state. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation.
Science, 345, 2014
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4V85
 
 | | Crystal Structure of Release Factor RF3 Trapped in the GTP State on a Rotated Conformation of the Ribosome. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Trakhanov, S, Noller, H.F. | | Deposit date: | 2011-06-13 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome.
Rna, 18, 2012
|
|
4UU9
 
 | | Crystal structure of the human c5a in complex with MEDI7814 a neutralising antibody | | Descriptor: | COMPLEMENT C5, MEDI7814, SULFATE ION | | Authors: | Colley, C, Sridharan, S, Dobson, C, Popovic, B, Debreczeni, J.E, Hargreaves, D, Edwards, B, Brennan, J, England, L, Fung, S, An Eghobamien, L, Sivars, U, Woods, R, Flavell, L, Renshaw, G.J, Wickson, K, Wilkinson, T, Davies, R, Bonnell, J, Warrener, P, Howes, R, Vaughan, T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors.
MAbs, 10, 2018
|
|
4V9M
 
 | | 70S Ribosome translocation intermediate FA-4.2A containing elongation factor EFG/FUSIDIC ACID/GDP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
4V7B
 
 | | Visualization of two tRNAs trapped in transit during EF-G-mediated translocation | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Lancaster, L, Sprink, T, Mielke, T, Loerke, J, Noller, H.F, Spahn, C.M.T. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V9K
 
 | | 70S ribosome translocation intermediate GDPNP-I containing elongation factor EFG/GDPNP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-24 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
4V9L
 
 | | 70S Ribosome translocation intermediate FA-3.6A containing elongation factor EFG/FUSIDIC ACID/GDP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-24 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
1XSM
 
 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
5T71
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with SA-2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-diazenyl]benzene-1-sulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5T74
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,5-bis(oxidanylidene)pyrrol-1-yl]-~{N}-(4-sulfamoylphenyl)ethanamide, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5T75
 
 | | Human carbonic anhydrase II G132C_C206S double mutant in complex with SA-2 | | Descriptor: | 4-[(E)-(4-aminophenyl)diazenyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5T72
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-(4-aminophenyl)diazenyl]benzenesulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5A0A
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-[(6R)-5-ethanoyl-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidin-6-yl]pyridine-2-carbonitrile, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
5A0C
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | (6S)-6-(4-cyano-2-methylsulfonyl-phenyl)-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-5-carbonitrile, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
5A09
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-[(4R)-4-(4-cyanophenyl)-5-ethanoyl-6-methyl-2-oxidanylidene-1-[3-(trifluoromethyl)phenyl]-4H-pyrimidin-3-yl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUTROPHIL ELASTASE, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
1XHX
 
 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1XHZ
 
 | | Phi29 DNA polymerase, orthorhombic crystal form, ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*T)-3', DNA polymerase | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1MY5
 
 | | NF-kappaB p65 subunit dimerization domain homodimer | | Descriptor: | NF-kappaB p65 (RelA) subunit | | Authors: | Huxford, T, Mishler, D, Phelps, C.B, Huang, D.-B, Sengchanthalangsy, L.L, Reeves, R, Hughes, C.A, Komives, E.A, Ghosh, G. | | Deposit date: | 2002-10-03 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Solvent exposed non-contacting amino acids play a critical role in NF-kappaB/I kappaB alpha complex formation
J.Mol.Biol., 324, 2002
|
|
1MY7
 
 | | NF-kappaB p65 subunit dimerization domain homodimer N202R mutation | | Descriptor: | NF-kappaB p65 (RelA) subunit | | Authors: | Huxford, T, Mishler, D, Phelps, C.B, Huang, D.-B, Sengchanthalangsy, L.L, Reeves, R, Hughes, C.A, Komives, E.A, Ghosh, G. | | Deposit date: | 2002-10-03 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Solvent exposed non-contacting amino acids play a critical role in NF-kappaB/IkappaB alpha complex formation
J.Mol.Biol., 324, 2002
|
|
7QJQ
 
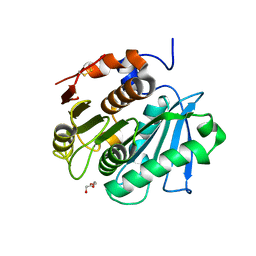 | | Crystal structure of a cutinase enzyme from Thermobifida fusca NTU22 (702) | | Descriptor: | Acetylxylan esterase, DI(HYDROXYETHYL)ETHER | | Authors: | Zahn, M, Gill, R.S, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
