6E0X
 
 | |
5WOF
 
 | | 1.65 ANGSTROM STRUCTURE OF THE DYNEIN LIGHT CHAIN 1 FROM PLASMODIUM FALCIPARUM | | Descriptor: | Dynein light chain 1, putative | | Authors: | Walker, J.R, Lew, J, Amani, M, Alam, Z, Wasney, G, Boulanger, K, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Botchkarev, A, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genome-scale Protein Expression and Structural Biology of
Plasmodium Falciparum and Related Apicomplexan Organisms.
MOL.BIOCHEM.PARASITOL., 151, 2007
|
|
6AMG
 
 | | cyt P460 of Nitrosomonas sp. AL212 | | Descriptor: | Cytochrome P460, HEME C | | Authors: | Smith, M, Lancaster, K. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Eponymous Cofactors in Cytochrome P460s from Ammonia-Oxidizing Bacteria Are Iron Porphyrinoids Whose Macrocycles Are Dibasic.
Biochemistry, 57, 2018
|
|
2A99
 
 | | Crystal structure of recombinant chicken sulfite oxidase at resting state | | Descriptor: | CHLORIDE ION, GLYCEROL, MOLYBDENUM ATOM, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A9A
 
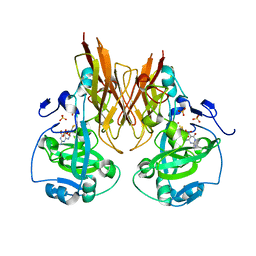 | | Crystal structure of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFATE ION, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2HU2
 
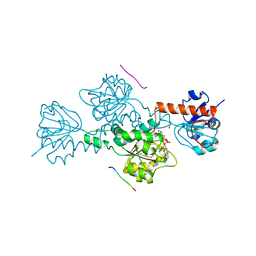 | | CTBP/BARS in ternary complex with NAD(H) and RRTGAPPAL peptide | | Descriptor: | 9-mer peptide from Zinc finger protein 217, C-terminal-binding protein 1, FORMIC ACID, ... | | Authors: | Nardini, M, Bolognesi, M, Quinlan, K.G.R, Verger, A, Francescato, P, Crossley, M. | | Deposit date: | 2006-07-26 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Specific Recognition of ZNF217 and Other Zinc Finger Proteins at a Surface Groove of C-Terminal Binding Proteins
Mol.Cell.Biol., 26, 2006
|
|
6UVU
 
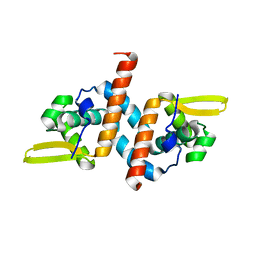 | | Crystal structure of the AntR antimony-specific transcriptional repressor | | Descriptor: | ArsR family transcriptional regulator | | Authors: | Thiruselvam, V, Banumathi, S, Palani, K, Manohar, R, Rosen, B.P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of AntR, an Sb(III) responsive transcriptional repressor.
Mol.Microbiol., 116, 2021
|
|
4JPH
 
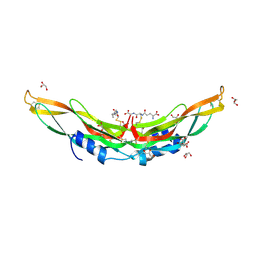 | | Crystal structure of Protein Related to DAN and Cerberus (PRDC) | | Descriptor: | CITRIC ACID, GLUTATHIONE, GLYCEROL, ... | | Authors: | Deng, X, Nolan, K.T, Kattamuri, C, Thompson, T.B. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein related to dan and cerberus: insights into the mechanism of bone morphogenetic protein antagonism.
Structure, 21, 2013
|
|
3C5D
 
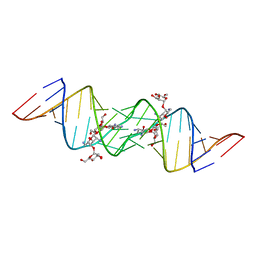 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to lividomycin | | Descriptor: | 'HIV-1 subtype F genomic RNA, (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, POTASSIUM ION | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3C7R
 
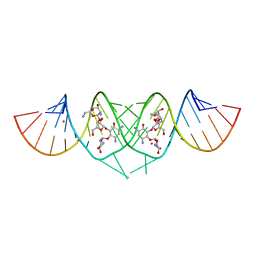 | | Crystal Structure of HIV-1 subtype F DIS extended duplex RNA bound to neomycin | | Descriptor: | HIV-1 subtype F genomic RNA, NEOMYCIN, POTASSIUM ION | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3TE3
 
 | |
4LAR
 
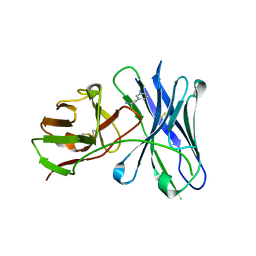 | | Crystal structure of a therapeutic single chain antibody in complex with amphetamine | | Descriptor: | (2S)-1-phenylpropan-2-amine, Single chain antibody fragment scFv6H4 | | Authors: | Celical, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
4LAQ
 
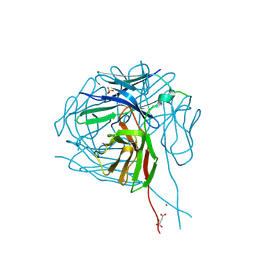 | | Crystal structure of a therapeutic single chain antibody in the free form | | Descriptor: | D-MALATE, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Celikel, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
6DCS
 
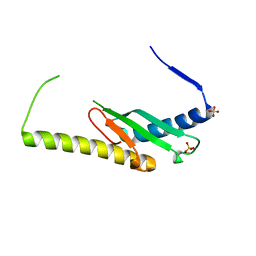 | | Stage III sporulation protein AF (SpoIIIAF) | | Descriptor: | SULFATE ION, Stage III sporulation protein AF | | Authors: | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | Deposit date: | 2018-05-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRY
 
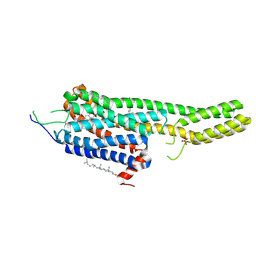 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4LAS
 
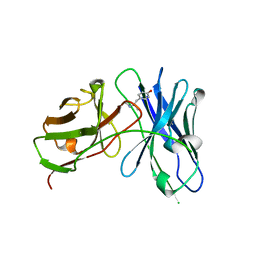 | | Crystal structure of a therapeutic single chain antibody in complex with 4-hydroxymethamphetamine | | Descriptor: | 4-[(2S)-2-(methylamino)propyl]phenol, Single chain antibody fragment scFv6H4 | | Authors: | Celical, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
3EII
 
 | | Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-16 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
3EJA
 
 | | Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
3C3Z
 
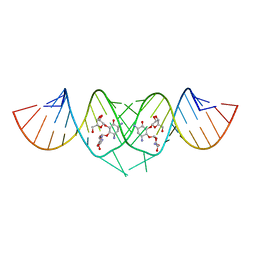 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to ribostamycin | | Descriptor: | HIV-1 subtype F genomic RNA, RIBOSTAMYCIN | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
7K32
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K33
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K30
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | Descriptor: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
