1ZS9
 
 | | Crystal structure of human enolase-phosphatase E1 | | Descriptor: | E-1 ENZYME, MAGNESIUM ION | | Authors: | Wang, H, Pang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-23 | | Release date: | 2005-06-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human E1 Enzyme and its Complex with a Substrate Analog Reveals the Mechanism of its Phosphatase/Enolase
J.Mol.Biol., 348, 2005
|
|
6BOW
 
 | |
1ZOY
 
 | | Crystal Structure of Mitochondrial Respiratory Complex II from porcine heart at 2.4 Angstroms | | Descriptor: | FAD-binding protein, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-15 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZAF
 
 | | Crystal structure of estrogen receptor beta complexed with 3-Bromo-6-hydroxy-2-(4-hydroxy-phenyl)-inden-1-one | | Descriptor: | 3-BROMO-6-HYDROXY-2-(4-HYDROXYPHENYL)-1H-INDEN-1-ONE, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | McDevitt, R.E, Malamas, M.S, Manas, E.S, Unwalla, R.J, Xu, Z.B, Miller, C.P, Harris, H.A. | | Deposit date: | 2005-04-06 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Estrogen receptor ligands: design and synthesis of new 2-arylindene-1-ones
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6BOV
 
 | |
6BOQ
 
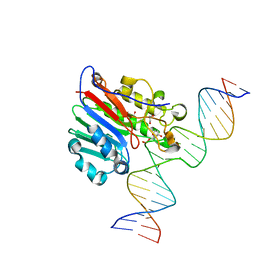 | | Human APE1 substrate complex with an A/A mismatch adjacent the THF | | Descriptor: | 1,2-ETHANEDIOL, 21-mer DNA, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BOU
 
 | |
6IKK
 
 | |
6JC4
 
 | |
5WQN
 
 | |
6BOS
 
 | |
6IN8
 
 | | Crystal structure of MucB | | Descriptor: | Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6JYV
 
 | | Structure of an isopenicillin N synthase from Pseudomonas aeruginosa PAO1 | | Descriptor: | Probable iron/ascorbate oxidoreductase, SODIUM ION | | Authors: | Hao, Z, Che, S, Wang, R, Liu, R, Zhang, Q, Bartlam, M. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural characterization of an isopenicillin N synthase family oxygenase from Pseudomonas aeruginosa PAO1.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
2BHW
 
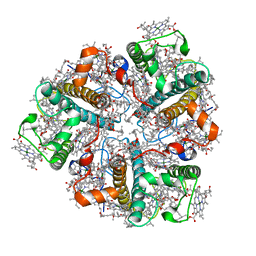 | | PEA LIGHT-HARVESTING COMPLEX II AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6'S,9R,9'R,13R,13'S)-4',5'-DIDEHYDRO-5',6',7',8',9,9',10,10',11,11',12,12',13,13',14,14',15,15'-OCTADECAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Standfuss, J, Terwisscha van Scheltinga, A.C, Lamborghini, M, Kuehlbrandt, W. | | Deposit date: | 2005-01-19 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of Photoprotection and Nonphotochemical Quenching in Pea Light-Harvesting Complex at 2.5 A Resolution.
Embo J., 24, 2005
|
|
2AMQ
 
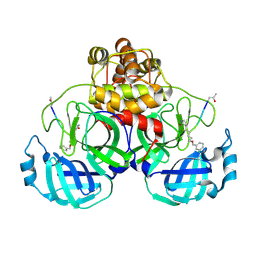 | | Crystal Structure Of SARS_CoV Mpro in Complex with an Inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Xue, X, Yang, K, Zhao, Q, Bartlam, M, Rao, Z. | | Deposit date: | 2005-08-10 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
5WQM
 
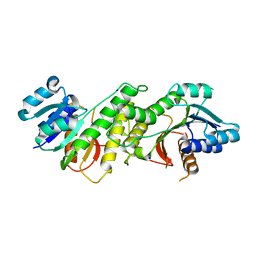 | |
2AHM
 
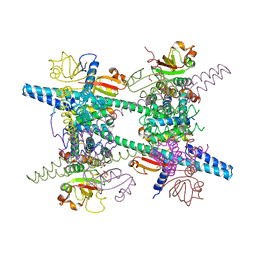 | | Crystal structure of SARS-CoV super complex of non-structural proteins: the hexadecamer | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, heavy chain, ... | | Authors: | Zhai, Y.J, Sun, F, Bartlam, M, Rao, Z. | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
6INB
 
 | |
6IKJ
 
 | | Crystal structure of YfiB(F48S) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
5WQO
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WQP
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
2AMP
 
 | | Crystal Structure Of Porcine Transmissible Gastroenteritis Virus Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Xue, X, Yang, K, Zhao, Q, Bartlam, M, Rao, Z. | | Deposit date: | 2005-08-10 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
2D2D
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor I2 | | Descriptor: | 3C-like proteinase, ETHYL (2E,4S)-4-[((2R)-2-{[N-(TERT-BUTOXYCARBONYL)-L-VALYL]AMINO}-2-PHENYLETHANOYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENT-2-ENOATE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Ding, Y, Rao, Z. | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
9GKY
 
 | | Crystal Structure of Histone deacetylase (HdaH) from Vibrio cholerae in complex with decanoic acid | | Descriptor: | DECANOIC ACID, Histone deacetylase, IMIDAZOLE, ... | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria
Nature Communications, 2024
|
|
9GKV
 
 | | Crystal Structure of Deacetylase (HdaH) from Vibrio cholerae in complex with SAHA | | Descriptor: | ACETATE ION, Histone deacetylase, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Graf, L.G, Schulze, S, Lammers, M, Palm, G.J. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria
Nature Communications, 2024
|
|
