2EVH
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mA7G) | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*CP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*GP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2EVI
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mA8T) | | Descriptor: | 5'-D(*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*TP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2EUX
 
 | | Structure of a Ndt80-DNA complex (MSE VARIANT vA4G) | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*GP*CP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*GP*CP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
5I2A
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diol-dehydratase | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
2EUZ
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mC5T) | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*AP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*TP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2EUW
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mA4T) | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*GP*AP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*TP*CP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
5I2G
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | Diol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
5I3T
 
 | | Native Structure of the Linalool Dehydratase-Isomerase from Castellaniella defragrans | | Descriptor: | 1,3-BUTANEDIOL, CHLORIDE ION, Linalool dehydratase/isomerase, ... | | Authors: | LaMattina, J.W, Carlock, M, Koch, D.J, Lanzilotta, W.N. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native Structure of the Linalool Dehydratase-Isomerase from Castellaniella defragrans
To Be Published
|
|
2EUV
 
 | |
2ETW
 
 | |
2EVJ
 
 | | Structure of an Ndt80-DNA complex (MSE mutant mA9C) | | Descriptor: | 5'-D(*AP*GP*TP*GP*TP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*AP*CP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2EVG
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mA7T) | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*AP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*TP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2EVF
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mA6T) | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*AP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*TP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
5FFQ
 
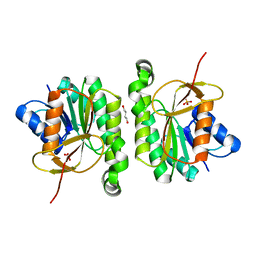 | | ChuY: An Anaerobillin Reductase from Escherichia coli O157:H7 | | Descriptor: | 1,4-BUTANEDIOL, PHOSPHATE ION, ShuY-like protein | | Authors: | LaMattina, J.W, Reedy, A.N, Uy, K.G, Lanzilotta, W.N. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Radical new paradigm for heme degradation in Escherichia coli O157:H7.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4MTJ
 
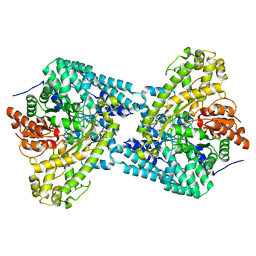 | | Structure of the b12-independent glycerol dehydratase with 1,2-propanediol bound | | Descriptor: | B12-independent glycerol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J, Wright, A.V, Demick, J, Soucaille, P, Lanzilotta, W.N. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | When Computational Chemistry and Modern Software Get It Right; New Insight Into the Mechanism of a Glycyl Radical Enzyme
To be Published
|
|
4QGS
 
 | |
1MN4
 
 | | Structure of Ndt80 (Residues 59-340) DNA-binding domain core | | Descriptor: | NDT80 PROTEIN | | Authors: | Lamoureux, J.S, Stuart, D, Tsang, R, Wu, C, Glover, J.N.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the sporulation-specific transcription factor Ndt80 bound to DNA
Embo J., 21, 2002
|
|
1MNN
 
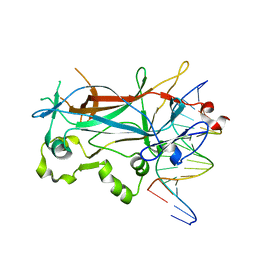 | | Structure of the sporulation specific transcription factor Ndt80 bound to DNA | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Stuart, D, Tsang, R, Wu, C, Glover, J.N. | | Deposit date: | 2002-09-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the sporulation-specific transcription factor Ndt80 bound to DNA
Embo J., 21, 2002
|
|
1OZU
 
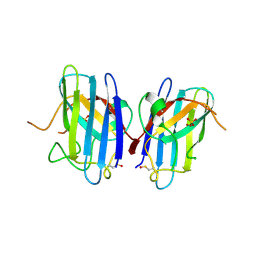 | | Crystal Structure of Familial ALS Mutant S134N of human Cu,Zn Superoxide Dismutase (CuZnSOD) to 1.3A resolution | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1P1V
 
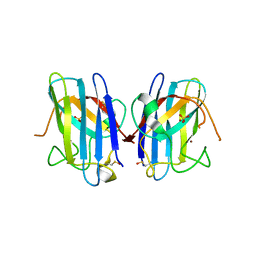 | | Crystal Structure of FALS-associated human Copper-Zinc Superoxide Dismutase (CuZnSOD) Mutant D125H to 1.4A | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Malek, K, Rodriguez, J.A, Doucette, P.A, Taylor, A.B, Hayward, L.J, Cabelli, D.E, Valentine, J.S, Hart, P.J. | | Deposit date: | 2003-04-14 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Alternative Mechanism of Bicarbonate-mediated Peroxidation by Copper-Zinc Superoxide Dismutase: RATES ENHANCED VIA PROPOSED ENZYME-ASSOCIATED PEROXYCARBONATE INTERMEDIATE
J.Biol.Chem., 278, 2003
|
|
1OZT
 
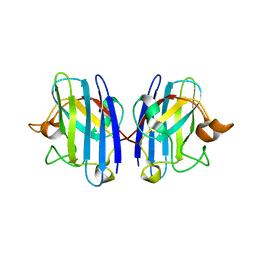 | | Crystal Structure of apo-H46R Familial ALS Mutant human Cu,Zn Superoxide Dismutase (CuZnSOD) to 2.5A resolution | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G2V
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G23
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-16 | | Release date: | 2000-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
