4R04
 
 | |
6OKR
 
 | |
1SHT
 
 | | Crystal Structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor | | Descriptor: | ACETATE ION, Anthrax toxin receptor 2, MAGNESIUM ION | | Authors: | Lacy, D.B, Wigelsworth, D.J, Scobie, H.M, Young, J.A.T, Collier, R.J. | | Deposit date: | 2004-02-26 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SHU
 
 | | Crystal Structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor | | Descriptor: | Anthrax toxin receptor 2, MAGNESIUM ION | | Authors: | Lacy, D.B, Wigelsworth, D.J, Scobie, H.M, Young, J.A.T, Collier, R.J. | | Deposit date: | 2004-02-26 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TZN
 
 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore bound to the VWA domain of CMG2, an anthrax toxin receptor | | Descriptor: | Anthrax toxin receptor 2, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | Deposit date: | 2004-07-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TZO
 
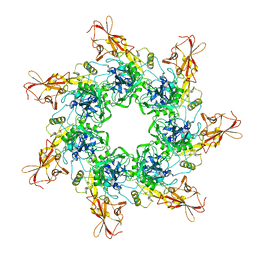 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | Deposit date: | 2004-07-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6O2N
 
 | |
6O2M
 
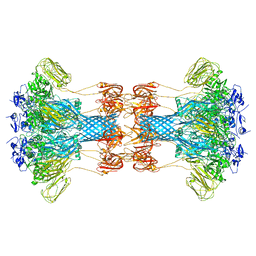 | |
6OKU
 
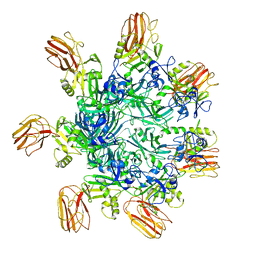 | |
6OKS
 
 | |
6O2O
 
 | |
6OKT
 
 | |
3SS1
 
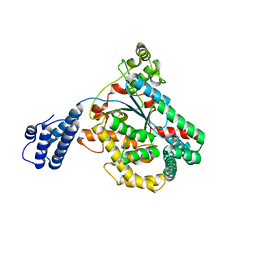 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain | | Descriptor: | MANGANESE (II) ION, Toxin A | | Authors: | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2011-07-07 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
3SRZ
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain bound to UDP-glucose | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2011-07-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
6V1S
 
 | | Structure of the Clostridioides difficile transferase toxin | | Descriptor: | ADP-ribosylating binary toxin enzymatic subunit CdtA, ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Sheedlo, M.J, Anderson, D.M, Thomas, A.K, Lacy, D.B. | | Deposit date: | 2019-11-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural elucidation of theClostridioides difficiletransferase toxin reveals a single-site binding mode for the enzyme.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3MPP
 
 | |
1J7N
 
 | | Anthrax Toxin Lethal factor | | Descriptor: | Lethal Factor precursor, SULFATE ION, ZINC ION | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-05-17 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
1JKY
 
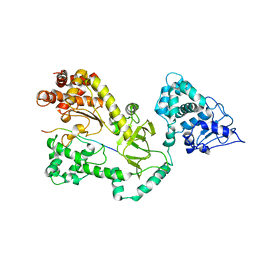 | | Crystal Structure of the Anthrax Lethal Factor (LF): Wild-type LF Complexed with the N-terminal Sequence of MAPKK2 | | Descriptor: | Lethal Factor, mitogen-activated protein kinase kinase 2 | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
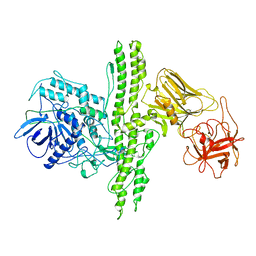
3BTA
 
 | |
6OEE
 
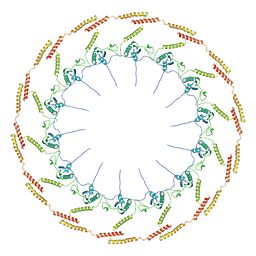 | | Structure of CagT from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagT | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagY | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
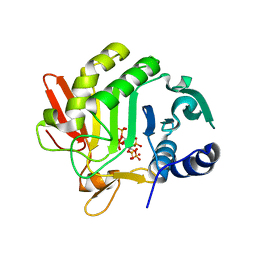
3HO6
 
 | |
7MUV
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 3 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUQ
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 1 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUY
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 5 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
