6O1L
 
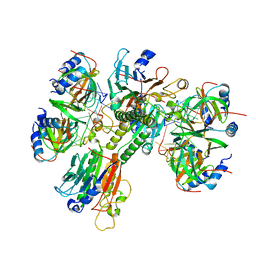 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression Hfq-Crc-amiE 2:3:2 complex | | 分子名称: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | 著者 | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | 登録日 | 2019-02-20 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Architectural principles for Hfq/Crc-mediated regulation of gene expression.
Elife, 8, 2019
|
|
6O1K
 
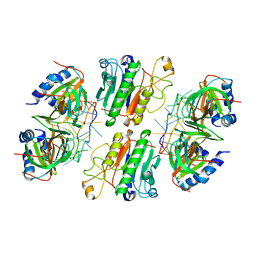 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression. Hfq-Crc-amiE 2:2:2 complex (core complex) | | 分子名称: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | 著者 | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | 登録日 | 2019-02-20 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Architectural principles for Hfq/Crc-mediated regulation of gene expression
Elife, 8, 2019
|
|
6O1M
 
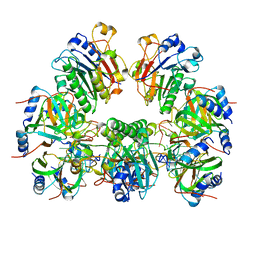 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression. Hfq-Crc-amiE 2:4:2 complex | | 分子名称: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | 著者 | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | 登録日 | 2019-02-20 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Architectural principles for Hfq/Crc-mediated regulation of gene expression.
Elife, 8, 2019
|
|
2YJN
 
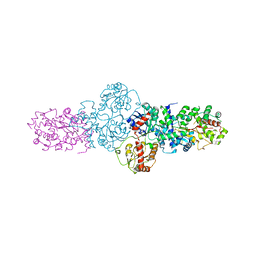 | | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII | | 分子名称: | DTDP-4-KETO-6-DEOXY-HEXOSE 3,4-ISOMERASE, GLYCOSYLTRANSFERASE | | 著者 | Moncrieffe, M.C, Fernandez, M.J, Spiteller, D, Matsumura, H, Gay, N.J, Luisi, B.F, Leadlay, P.F. | | 登録日 | 2011-05-20 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.091 Å) | | 主引用文献 | Structure of the Glycosyltransferase Eryciii in Complex with its Activating P450 Homologue Erycii.
J.Mol.Biol., 415, 2012
|
|
3H8A
 
 | |
4OWG
 
 | |
4O7J
 
 | |
5NC5
 
 | |
2YJV
 
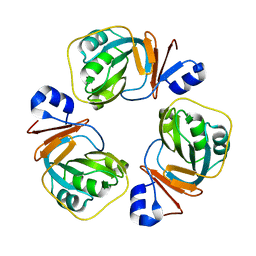 | |
352D
 
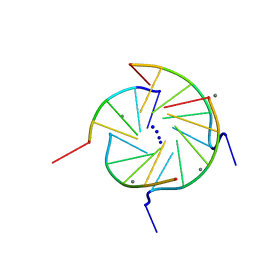 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | 分子名称: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | 著者 | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | 登録日 | 1997-09-04 | | 公開日 | 1997-11-10 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
2YJT
 
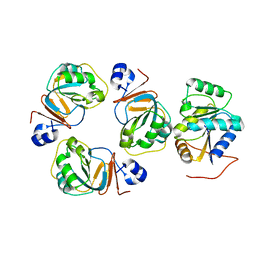 | |
6TNN
 
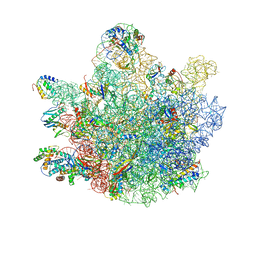 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | 分子名称: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | 登録日 | 2019-12-09 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6TPQ
 
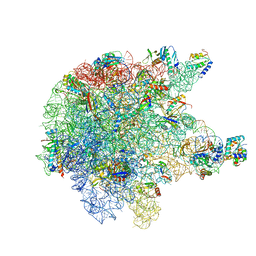 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | 分子名称: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | 登録日 | 2019-12-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
4QKK
 
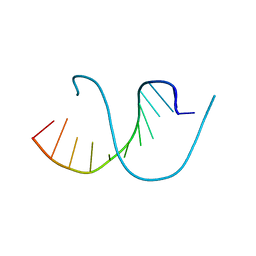 | | Crystal structure of an oligonucleotide containing 5-formylcytosine | | 分子名称: | DNA (5'-D(*CP*TP*AP*(5FC)P*GP*(5FC)P*GP*(5FC)P*GP*TP*AP*G)-3') | | 著者 | Raiber, E.-A, Murat, P, Chirgadze, D.Y, Luisi, B.F, Balasubramanian, S. | | 登録日 | 2014-06-06 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | 5-Formylcytosine alters the structure of the DNA double helix.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8BVJ
 
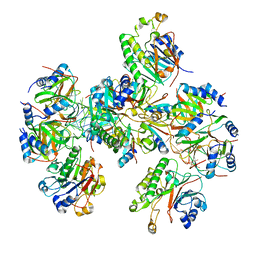 | | Hfq-Crc-estA translation repression complex | | 分子名称: | Catabolite repression control protein, RNA-binding protein Hfq, estA mRNA | | 著者 | Dendooven, T, Luisi, B.F. | | 登録日 | 2022-12-04 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Translational regulation by Hfq-Crc assemblies emerges from polymorphic ribonucleoprotein folding.
Embo J., 42, 2023
|
|
4C48
 
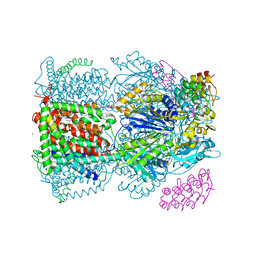 | | Crystal structure of AcrB-AcrZ complex | | 分子名称: | ACRIFLAVINE RESISTANCE PROTEIN B, DARPIN, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Du, D, James, N, Klimont, E, Luisi, B.F. | | 登録日 | 2013-09-02 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of the AcrAB-TolC multidrug efflux pump.
Nature, 509, 2014
|
|
4AM3
 
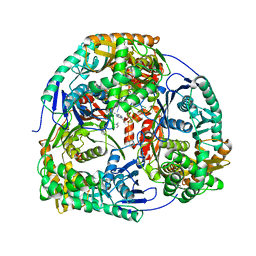 | | Crystal structure of C. crescentus PNPase bound to RNA | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-03-07 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
3CFS
 
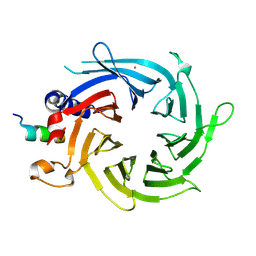 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | 分子名称: | ARSENIC, Histone H4, Histone-binding protein RBBP7 | | 著者 | Murzina, N.V, Pei, X.-Y, Pratap, J.V, Sparkes, M, Vicente-Garcia, J, Ben-Shahar, T.R, Verreault, A, Luisi, B.F, Laue, E.D. | | 登録日 | 2008-03-04 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
4AID
 
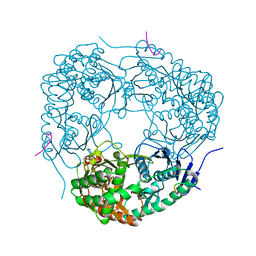 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-02-09 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
3CFV
 
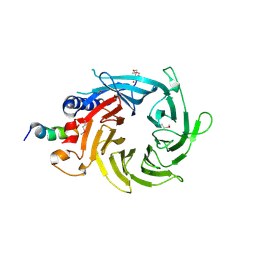 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | 分子名称: | ARSENIC, Histone H4 peptide, Histone-binding protein RBBP7 | | 著者 | Pei, X.-Y, Murzina, N.V, Zhang, W, McLaughlin, S, Verreault, A, Luisi, B.F, Laue, E.D. | | 登録日 | 2008-03-04 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
4CDI
 
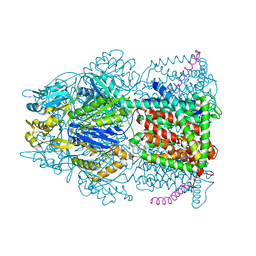 | | Crystal structure of AcrB-AcrZ complex | | 分子名称: | ACRIFLAVINE RESISTANCE PROTEIN B, PREDICTED PROTEIN | | 著者 | Du, D, James, N, Klimont, E, Luisi, B.F. | | 登録日 | 2013-10-31 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structure of the Acrab-Tolc Multidrug Efflux Pump.
Nature, 509, 2014
|
|
1EK9
 
 | | 2.1A X-RAY STRUCTURE OF TOLC: AN INTEGRAL OUTER MEMBRANE PROTEIN AND EFFLUX PUMP COMPONENT FROM ESCHERICHIA COLI | | 分子名称: | OUTER MEMBRANE PROTEIN TOLC | | 著者 | Koronakis, V, Sharff, A.J, Koronakis, E, Luisi, B, Hughes, C. | | 登録日 | 2000-03-07 | | 公開日 | 2000-06-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export.
Nature, 405, 2000
|
|
4AIM
 
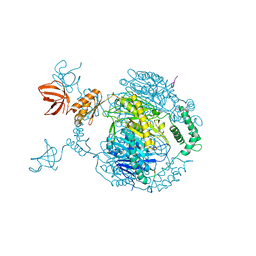 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-02-10 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
1W85
 
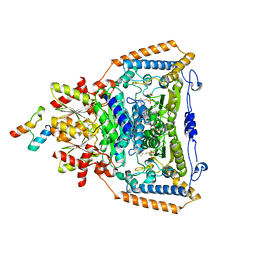 | | The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2 | | 分子名称: | DI(HYDROXYETHYL)ETHER, DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, ... | | 著者 | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | 登録日 | 2004-09-16 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A molecular switch and proton wire synchronize the active sites in thiamine enzymes.
Science, 306, 2004
|
|
1W88
 
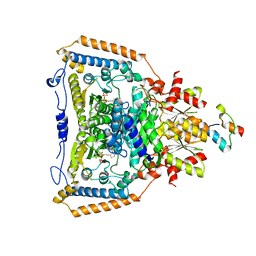 | | The crystal structure of pyruvate dehydrogenase E1(D180N,E183Q) bound to the peripheral subunit binding domain of E2 | | 分子名称: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, PYRUVATE DEHYDROGENASE E1 COMPONENT, ... | | 著者 | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | 登録日 | 2004-09-16 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Molecular Switch and Proton-Wire Synchronize the Active Sites in Thiamine-Dependent Enzymes
Science, 306, 2004
|
|
