6LBO
 
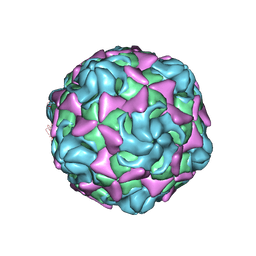 | | Cryo-EM structure of echovirus 11 empty particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LBQ
 
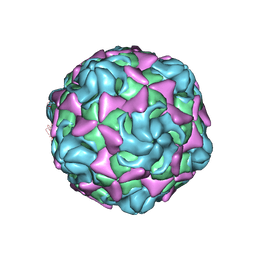 | | Cryo-EM structure of echovirus 11 empty particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LA4
 
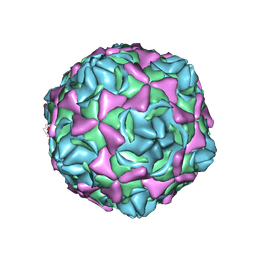 | | Cryo-EM structure of full echovirus 11 particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LA6
 
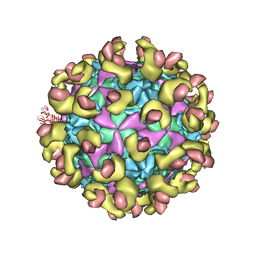 | |
6LAP
 
 | | Cryo-EM structure of echovirus 11 A-particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6M1K
 
 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
5BX1
 
 | | Crystal Structure of PRL-1 complex with compound analogy 3 | | Descriptor: | 3-(5,6-dimethyl-2H-isoindol-2-yl)-N'-[(E)-furan-2-ylmethylidene]benzohydrazide, Protein tyrosine phosphatase type IVA 1, SULFATE ION | | Authors: | Liu, S, Bai, Y, Zhang, Z. | | Deposit date: | 2015-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of PRL-1 complex with compound analogy 3
To Be Published
|
|
8G93
 
 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G84
 
 | | Crystal structures of HSD17B13 complexes | | Descriptor: | Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G89
 
 | | HSD17B13 in complex with cofactor and inhibitor | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
4KBK
 
 | |
4KBA
 
 | | CK1d in complex with 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine inhibitor | | Descriptor: | 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Liu, S. | | Deposit date: | 2013-04-23 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ligand-protein interactions of selective casein kinase 1 delta inhibitors.
J.Med.Chem., 56, 2013
|
|
5HHX
 
 | |
4KB8
 
 | |
2GV9
 
 | |
5HHV
 
 | |
4KBC
 
 | |
2I87
 
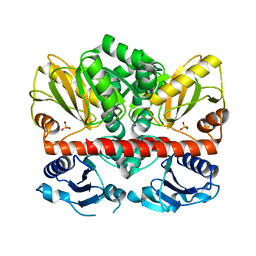 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | D-alanine-D-alanine ligase, SULFATE ION | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I8C
 
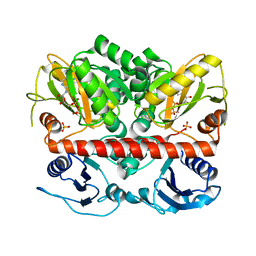 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I80
 
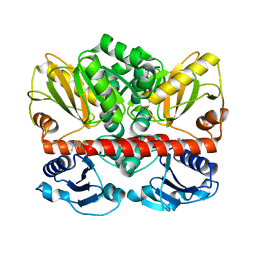 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | 3-CHLORO-2,2-DIMETHYL-N-[4-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE, D-alanine-D-alanine ligase | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M.-M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4HR9
 
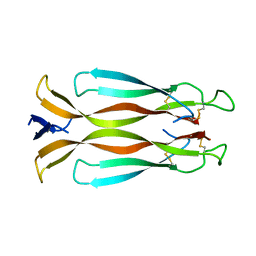 | | Human interleukin 17A | | Descriptor: | Interleukin-17A | | Authors: | Liu, S. | | Deposit date: | 2012-10-26 | | Release date: | 2013-05-22 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structures of interleukin 17A and its complex with IL-17 receptor A.
Nat Commun, 4, 2013
|
|
4HSA
 
 | | Structure of interleukin 17a in complex with il17ra receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Liu, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structures of interleukin 17A and its complex with IL-17 receptor A.
Nat Commun, 4, 2013
|
|
7UOM
 
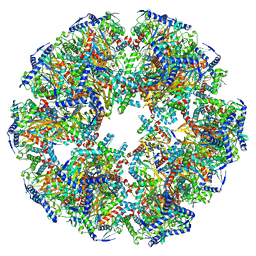 | | Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruvate dehydrogenase complex from bovine kidney | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Liu, S, Xia, X, Zhen, J, Li, Z.H, Zhou, Z.H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney.
Cell Discov, 8, 2022
|
|
7UOL
 
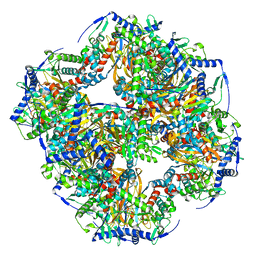 | | Endogenous dihydrolipoamide succinyltransferase (E2) core of 2-oxoglutarate dehydrogenase complex from bovine kidney | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Liu, S, Xia, X, Zhen, J, Li, Z.H, Zhou, Z.H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney.
Cell Discov, 8, 2022
|
|
4DHY
 
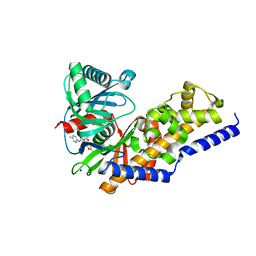 | | Crystal structure of human glucokinase in complex with glucose and activator | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
