6KLY
 
 | | Crystal structure of the type III effector XopAI from Xanthomonas axonopodis pv. citri in space group P43212 | | Descriptor: | Type III effector XopAI | | Authors: | Liu, J.-H, Wu, J.E, Lin, H, Chiu, S.W, Yang, J.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure-Based Exploration of Arginine-Containing Peptide Binding in the ADP-Ribosyltransferase Domain of the Type III Effector XopAI Protein.
Int J Mol Sci, 20, 2019
|
|
4LCY
 
 | | Crystal structure of HLA-b46 at 1.6 angstrom resolution | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-46 alpha chain, ... | | Authors: | Liu, J.X. | | Deposit date: | 2013-06-24 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure Analysis of the Human Leukocyte Antigen B*46:01
To be Published
|
|
6K94
 
 | |
3OX8
 
 | | Crystal Structure of HLA A*02:03 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
2LSJ
 
 | |
2LSG
 
 | |
3R00
 
 | |
6CI0
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with E101A (II) mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2018-02-23 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The K-path entrance in cytochrome c oxidase is defined by mutation of E101 and controlled by an adjacent ligand binding domain.
Biochim. Biophys. Acta, 1859, 2018
|
|
8XYS
 
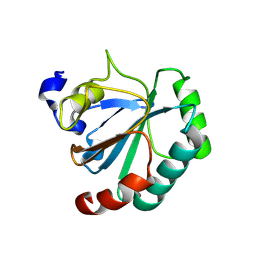 | | De novo designed protein GPX4-1 | | Descriptor: | De novo designed GPX4-1 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYW
 
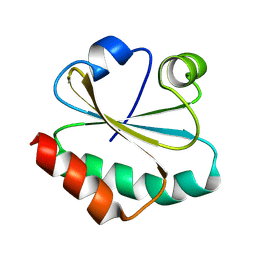 | | De novo designed protein Trx-3 | | Descriptor: | De novo designed Trx-3 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYT
 
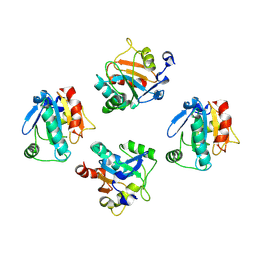 | | De novo designed protein GPX4-4 | | Descriptor: | De novo designed GPX4-4 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYV
 
 | | De novo designed protein 0705-5 | | Descriptor: | De novo designed protein 0705-5 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
3R01
 
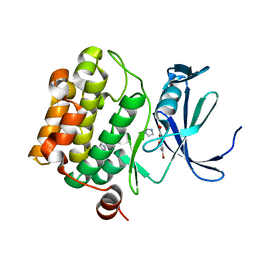 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-bromo-7-methoxy-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Liu, J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8I0H
 
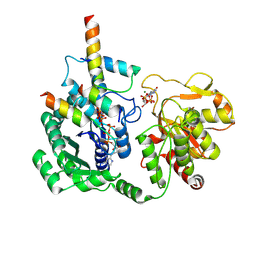 | |
2MTP
 
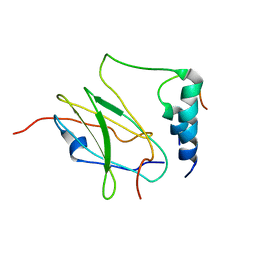 | |
2GUV
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
3OXS
 
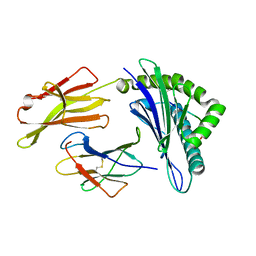 | | Crystal Structure of HLA A*02:07 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3U91
 
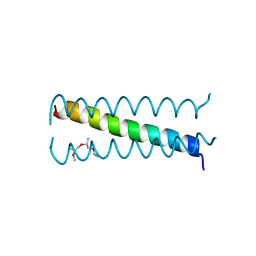 | |
3OXR
 
 | | Crystal Structure of HLA A*02:06 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
2GUS
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
2L3T
 
 | | Solution structure of tandem SH2 domain from Spt6 | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Liu, J, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-09-22 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tandem SH2 domains from Spt6 and their binding to the phosphorylated RNA polymerase II C-terminal domain
To be Published
|
|
7RAM
 
 | | Cryo-EM Structure of the HCMV gHgLgO Trimer Derived from AD169 and TR strains in complex with PDGFRalpha | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, Envelope glycoprotein O, ... | | Authors: | Liu, J, Vanarsdall, A.L, Chen, D, Johnson, D.C, Jardetzky, T.S. | | Deposit date: | 2021-07-02 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-Electron Microscopy Structure and Interactions of the Human Cytomegalovirus gHgLgO Trimer with Platelet-Derived Growth Factor Receptor Alpha.
Mbio, 12, 2021
|
|
4OK1
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OH5
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OHA
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
