4G43
 
 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | 著者 | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | 登録日 | 2012-07-16 | | 公開日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
5WWJ
 
 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | 登録日 | 2017-01-01 | | 公開日 | 2018-01-17 | | 最終更新日 | 2019-07-31 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1).
To Be Published
|
|
3ISW
 
 | | Crystal structure of filamin-A immunoglobulin-like repeat 21 bound to an N-terminal peptide of CFTR | | 分子名称: | Cystic fibrosis transmembrane conductance regulator, Filamin-A | | 著者 | Xu, Z, Page, R, Qin, J, Ithychanda, S.S, Liu, J.M, Misra, S. | | 登録日 | 2009-08-27 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin.
J.Biol.Chem., 285, 2010
|
|
1MJN
 
 | | Crystal Structure of the intermediate affinity aL I domain mutant | | 分子名称: | Integrin alpha-L, MAGNESIUM ION | | 著者 | Shimaoka, M, Xiao, T, Liu, J.H, Yang, Y.T, Dong, Y.C, Jun, C.D, McCormack, A, Zhang, R.G, Wang, J.H, Springer, T.A. | | 登録日 | 2002-08-28 | | 公開日 | 2003-01-28 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structures of the alphaL I Domain and its Complex with ICAM-1 reveal a Shape-shifting Pathway for Integrin Regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
2YP7
 
 | | Haemagglutinin of 2005 Human H3N2 Virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP5
 
 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Avian Receptor Analogue 3SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP2
 
 | | Haemagglutinin of 2004 Human H3N2 Virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4NDZ
 
 | | Structure of Maltose Binding Protein fusion to 2-O-Sulfotransferase with bound heptasaccharide and PAP | | 分子名称: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1 fusion, ... | | 著者 | Liu, C, Sheng, J, Krahn, J.M, Perera, L, Xu, Y, Hsieh, P, Liu, J, Pedersen, L.C. | | 登録日 | 2013-10-28 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Deciphering the role of 2-O-sulfotransferase in regulating heparan sulfate biosynthesis
To be Published
|
|
2YP8
 
 | | Haemagglutinin of 2005 Human H3N2 Virus in Complex with Human Receptor Analogue 6SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BGW
 
 | | Crystal Structure of H5 (VN1194) Influenza Haemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2013-03-29 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
1R6Z
 
 | | The Crystal Structure of the Argonaute2 PAZ domain (as a MBP fusion) | | 分子名称: | Chimera of Maltose-binding periplasmic protein and Argonaute 2, NICKEL (II) ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Song, J.J, Liu, J, Tolia, N.H, Schneiderman, J, Smith, S.K, Martienssen, R.A, Hannon, G.J, Joshua-Tor, L. | | 登録日 | 2003-10-17 | | 公開日 | 2004-01-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes.
Nat.Struct.Biol., 10, 2003
|
|
4B7N
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | 著者 | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2012-08-21 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4UC1
 
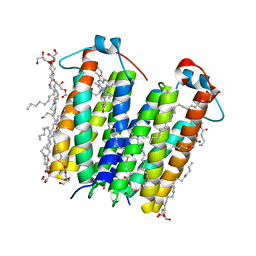 | | High resolution crystal structure of translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, METHOXY-ETHOXYL, ... | | 著者 | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | 登録日 | 2014-08-13 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
4UC2
 
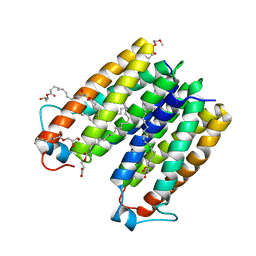 | | Crystal structure of translocator protein 18kDa (TSPO) from rhodobacter sphaeroides (A139T mutant) in P212121 space group | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, TETRAETHYLENE GLYCOL, TRANSLOCATOR PROTEIN TSPO | | 著者 | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | 登録日 | 2014-08-13 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
1OJ1
 
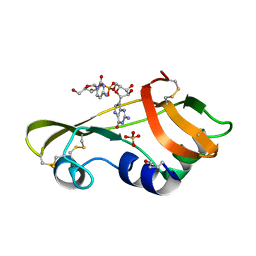 | | Nonproductive and Novel Binding Modes in Cytotoxic Ribonucleases from Rana catesbeiana of Two Crystal Structures Complexed with (2,5 CpG) and d(ApCpGpA) | | 分子名称: | CYTIDYL-2'-5'-PHOSPHO-GUANOSINE, RC-RNASE6 RIBONUCLEASE, SULFATE ION | | 著者 | Tsai, C.-J, Liu, J.-H, Liao, Y.-D, Chen, L.-y, Cheng, P.-T, Sun, Y.-J. | | 登録日 | 2003-06-28 | | 公開日 | 2004-07-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Nonproductive and Novel Binding Modes in Cytotoxic Ribonucleases from Rana Catesbeiana of Two Crystal Structures Complexed with C(2,5 Cpg) and D(Apcpgpa)
To be Published
|
|
4B7J
 
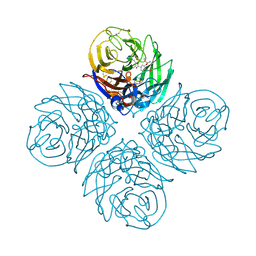 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | 分子名称: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2012-08-20 | | 公開日 | 2012-10-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.417 Å) | | 主引用文献 | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
2YP3
 
 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Human Receptor Analogue 6SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YP4
 
 | | Haemagglutinin of 2004 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | 著者 | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5WWI
 
 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | 登録日 | 2017-01-01 | | 公開日 | 2018-01-17 | | 最終更新日 | 2019-07-31 | | 実験手法 | X-RAY DIFFRACTION (3.194 Å) | | 主引用文献 | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1).
To Be Published
|
|
4BGY
 
 | | H5 (VN1194) Influenza Virus Haemagglutinin in Complex with Avian Receptor Analogue 3'-SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | 著者 | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2013-03-29 | | 公開日 | 2013-04-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
4BGX
 
 | | H5 (VN1194) Influenza Virus Haemagglutinin in Complex with Human Receptor Analogue 6'-SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | 著者 | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | 登録日 | 2013-03-29 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
1EQ7
 
 | |
6IN5
 
 | |
7NIH
 
 | | Wzc-K540M MgADP C8 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | 著者 | Naismith, J.H, Liu, J.W, Yang, Y. | | 登録日 | 2021-02-12 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NII
 
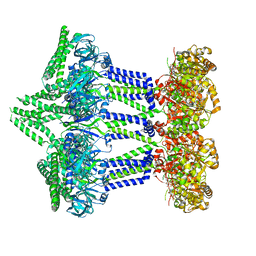 | | Wzc-K540M MgADP C1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | 著者 | Naismith, J.H, Liu, J.W, Yang, Y. | | 登録日 | 2021-02-12 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
