5H6G
 
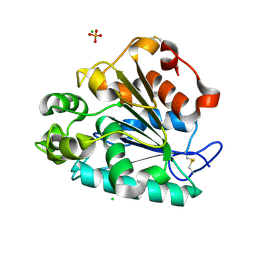 | | Crystal structure of a thermostable lipase from Marine Streptomyces | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hou, S, Zhao, Z, Liu, J. | | Deposit date: | 2016-11-11 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a lipase from Streptomyces sp. strain W007 - implications for thermostability and regiospecificity
FEBS J., 284, 2017
|
|
6JBW
 
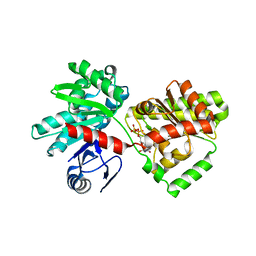 | | Structure of Tps1/UDP complex | | Descriptor: | Trehalose-6-phosphate synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
3DSG
 
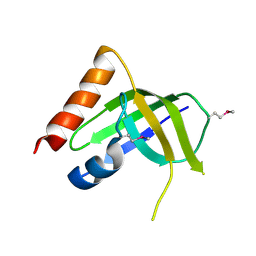 | | XC1028 from Xanthomonas campestris Adopts a PilZ Domain-like Structure Yet with Trivial c-di-GMP Binding Activity | | Descriptor: | Type IV fimbriae assembly protein | | Authors: | Li, T.N, Chin, K.H, Liu, J.H, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2008-07-12 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch.
Proteins, 75, 2009
|
|
7YOJ
 
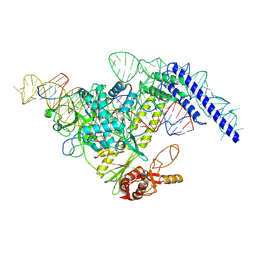 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
4LL9
 
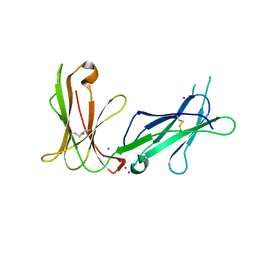 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
8I3X
 
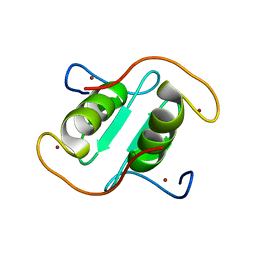 | | Rice APIP6-RING homodimer | | Descriptor: | RING-type domain-containing protein, ZINC ION | | Authors: | Zheng, Y, Zhang, X, Liu, Y, Liu, J, Wang, D. | | Deposit date: | 2023-01-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of rice APIP6 reveals a new dimerization mode of RING-type E3 ligases that facilities the construction of its working model
Phytopathol Res, 5, 2023
|
|
4HSV
 
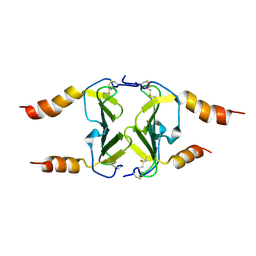 | | Crystal Structure of CXCL4L1 | | Descriptor: | Platelet factor 4 variant | | Authors: | Kuo, J.H, Liu, J.S, Wu, W.G, Sue, S.C. | | Deposit date: | 2012-10-30 | | Release date: | 2013-04-10 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Alternative C-terminal helix orientation alters chemokine function: structure of the anti-angiogenic chemokine, CXCL4L1
J.Biol.Chem., 288, 2013
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
7DV8
 
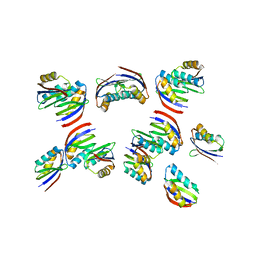 | | The crystal structure of rice immune receptor RGA5-HMA2. | | Descriptor: | Disease resistance protein RGA5 | | Authors: | Zhang, X, Liu, J.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4DN5
 
 | | Crystal Structure of NF-kB-inducing Kinase (NIK) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Liu, J, Sudom, A, Walker, N.P, Wang, Z. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nuclear Factor Kappa B-inducing kinase domain reveals a constitutively active conformation
J.Biol.Chem., 287, 2012
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
8GDX
 
 | |
8GE0
 
 | |
6C62
 
 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. | | Descriptor: | AtzG, Biuret hydrolase, MAGNESIUM ION | | Authors: | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | Deposit date: | 2018-01-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
6C6G
 
 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. Inhibitor bound complex. | | Descriptor: | AtzG, Biuret hydrolase, CALCIUM ION | | Authors: | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
6JCB
 
 | |
7VF5
 
 | | Human m6A-METTL associated complex (WTAP, VIRMA, and HAKAI) | | Descriptor: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog | | Authors: | Su, S, Li, S, Deng, T, Gao, M, Yin, Y, Wu, B, Peng, C, Liu, J, Ma, J, Zhang, K. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human m6A writer complexes.
Cell Res., 32, 2022
|
|
7VF2
 
 | | Human m6A-METTL associated complex (WTAP, VIRMA, ZC3H13, and HAKAI) | | Descriptor: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog, Zinc finger CCCH domain-containing protein 13 | | Authors: | Su, S, Li, S, Deng, T, Gao, M, Yin, Y, Wu, B, Peng, C, Liu, J, Ma, J, Zhang, K. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human m6A writer complexes.
Cell Res., 32, 2022
|
|
3AOF
 
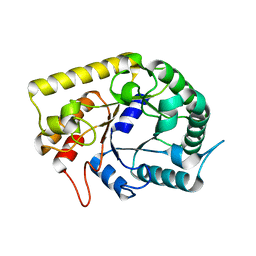 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
2KK1
 
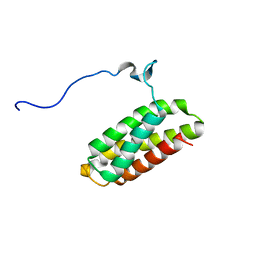 | | Solution structure of C-terminal Domain of Tyrosine-protein kinase ABL2 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR5537A | | Descriptor: | Tyrosine-protein kinase ABL2 | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-14 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of F-actin-binding domain of Arg/Abl2 from Homo sapiens.
Proteins, 78, 2010
|
|
4LLA
 
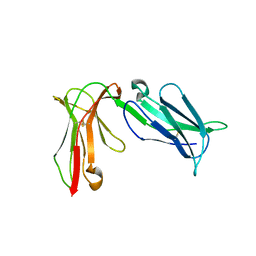 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
3AMD
 
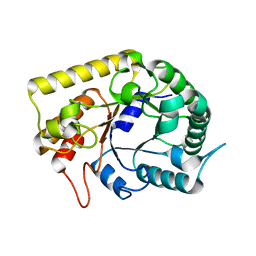 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
2KK0
 
 | | Solution structure of dead ringer-like protein 1 (at-rich interactive domain-containing protein 3a) from homo sapiens, northeast structural genomics consortium (NESG) target hr4394c | | Descriptor: | AT-rich interactive domain-containing protein 3A | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-14 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ARID domain of human AT-rich interactive domain-containing protein 3A: a human cancer protein interaction network target.
Proteins, 78, 2010
|
|
3AMC
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
