1QBY
 
 | | THE SOLUTION STRUCTURE OF A BAY-REGION 1R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*GP*GP*AP*CP*(BZA)AP*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP* G)-3' | | Authors: | Li, Z, Mao, H, Kim, H.-Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-04-27 | | Release date: | 1999-05-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (-)-(1R,2S,3R, 4S)-N6-[1-benz[a]anthracenyl]-2'-deoxyadenosyl adduct in an oligodeoxynucleotide containing the human N-ras codon 61 sequence.
Biochemistry, 38, 1999
|
|
2QC3
 
 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
8CWY
 
 | |
6U7H
 
 | | Cryo-EM structure of the HCoV-229E spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Z, Benlekbir, S, Rubinstein, J.L, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
8YEO
 
 | | Type I-FHNH Cascade-dsDNA R-loop complex | | Descriptor: | 60-nt crRNA, Cas5f, Cas6f, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YDB
 
 | | Type I-FHNH Cascade-dsDNA intermediate complex | | Descriptor: | 60-nt crRNA, Cas5f, Cas6f, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YB6
 
 | | Type I-EHNH Cascade complex | | Descriptor: | 61-nt crRNA, CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YH9
 
 | | Type I-FHNH Cascade complex | | Descriptor: | 60-nt crRNA, Cas5f, Cas6f, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8YHA
 
 | | Type I-EHNH Cascade-ssDNA complex | | Descriptor: | 61-nt crRNA, CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
7YFY
 
 | | Cryo-EM structure of the Mili-piRNA- target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (5'-R(P*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
8J9G
 
 | | CrtSPARTA hetero-dimer bound with guide-target, state 1 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J8H
 
 | | SPARTA monomer bound with guide-target, state 2 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J9P
 
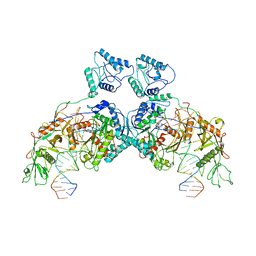 | | SPARTA dimer bound with guide-target | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
5JB1
 
 | | Pseudo-atomic structure of Human Papillomavirus Type 59 L1 Virus-like Particle | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Yan, X.D, Yu, H, Zheng, Q.B, Gu, Y, Li, S.W. | | Deposit date: | 2016-04-13 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
6LBE
 
 | |
5J6R
 
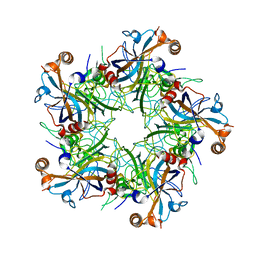 | | Crystal structure of Human Papillomavirus Type 59 L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Yan, X.D, Yu, H, Gu, Y, Li, S.W. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.011 Å) | | Cite: | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
5K08
 
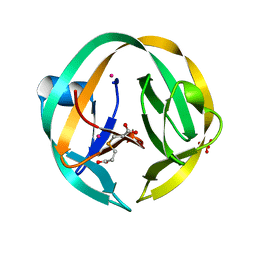 | | RecA mini intein-Zeise's salt complex | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, RecA mini intein, SULFATE ION, ... | | Authors: | Li, Z, Li, H.M. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
6MWY
 
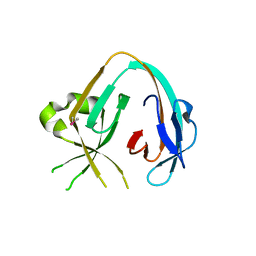 | | The Prp8 intein of Cryptococcus gattii | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Fu, B, Green, C.M, Lang, Y, Zhang, J, Oven, T.S, Li, X, Callahan, B.P, Chaturvedi, S, Belfort, M, Liao, G, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
6VSI
 
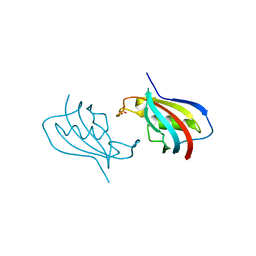 | | Crystal structure of FKBP12 of Candida auris | | Descriptor: | Peptidylprolyl isomerase, SULFATE ION | | Authors: | Li, Z, Li, H, Hernandez, G, LeMaster, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and transient dimerization for the FKBP12 protein from the pathogenic fungus Candida auris.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6MYL
 
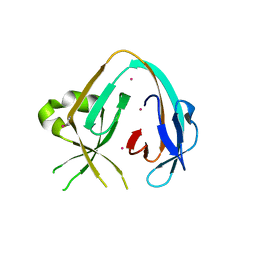 | | The Prp8 intein-cisplatin complex | | Descriptor: | PLATINUM (II) ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Li, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
6MX6
 
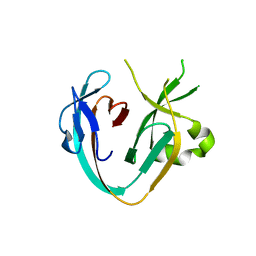 | | The Prp8 intein of Cryptococcus neoformans | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Spliceosomal Prp8 intein at the crossroads of protein and RNA splicing.
Plos Biol., 17, 2019
|
|
6OWU
 
 | |
7C23
 
 | | Crystal structure of CrmE10, a SGNH-hydrolase family esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7BQT
 
 | | Epstein-Barr virus, C12 portal dodecamer | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
6BS8
 
 | | The class 3 DnaB intein from Mycobacterium smegmatis | | Descriptor: | Replicative DNA helicase | | Authors: | Li, Z, Kelley, D.S, Banavali, N, Belfort, M, Li, H. | | Deposit date: | 2017-12-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mycobacterial DnaB helicase intein as oxidative stress sensor.
Nat Commun, 9, 2018
|
|
