4U0E
 
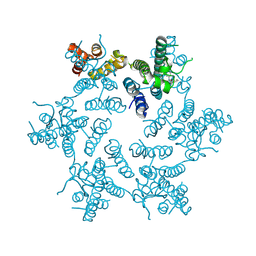 | | Hexameric HIV-1 CA in complex with PF3450074 | | Descriptor: | CHLORIDE ION, Capsid protein p24, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0B
 
 | |
6XUV
 
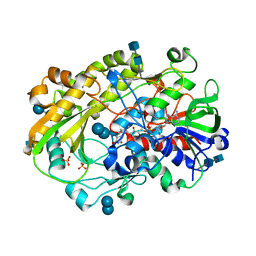 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, laminaribiose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
4U0F
 
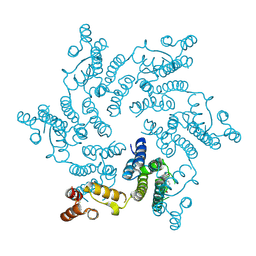 | | Hexameric HIV-1 CA in Complex with BI-2 | | Descriptor: | (4S)-4-(4-hydroxyphenyl)-3-phenyl-4,5-dihydropyrrolo[3,4-c]pyrazol-6(1H)-one, 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
6RA6
 
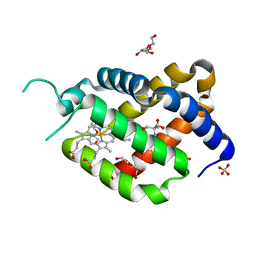 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
6TYU
 
 | |
4U0D
 
 | | Hexameric HIV-1 CA in complex with Nup153 peptide, P212121 crystal form | | Descriptor: | CHLORIDE ION, Gag polyprotein, Nuclear pore complex protein Nup153 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
6U7D
 
 | |
8SLR
 
 | | Crystal Structure of mouse TRAIL | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pedersen, L.C, Xu, D. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Heparan sulfate promotes TRAIL-induced tumor cell apoptosis.
Elife, 12, 2024
|
|
6XUU
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, glucose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6TYT
 
 | |
6XUT
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Sciara, G, Vallone, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6OKJ
 
 | |
6XRX
 
 | | Crystal structure of the mosquito protein AZ1 as an MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Foo, A.C.Y. | | Deposit date: | 2020-07-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mosquito protein AEG12 displays both cytolytic and antiviral properties via a common lipid transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8TCJ
 
 | |
8TCK
 
 | |
8TCL
 
 | |
8TCM
 
 | | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and Xanthene-1,3,6,8-tetrol bound | | Descriptor: | 9H-xanthene-1,3,6,8-tetrol, PICRIC ACID, p51 subunit | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and xanthene bound
To Be Published
|
|
6RXN
 
 | |
6XKG
 
 | | Crystal structure of 3-O-Sulfotransferase isoform 3 in complex with 8mer oligosaccharide with 6S sulfation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ... | | Authors: | Pedersen, L.C, Liu, J, Wander, R. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deciphering the substrate recognition mechanisms of the heparan sulfate 3- O -sulfotransferase-3.
Rsc Chem Biol, 2, 2021
|
|
6XL8
 
 | | Crystal structure of 3-O-Sulfotransferase isoform 3 in complex with 8mer oligosaccharide with no 6S sulfation | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 3A1, IODIDE ION, ... | | Authors: | Pedersen, L.C, Liu, J, Wander, R. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Deciphering the substrate recognition mechanisms of the heparan sulfate 3- O -sulfotransferase-3.
Rsc Chem Biol, 2, 2021
|
|
6VCJ
 
 | | Crystal structure of hsDHFR in complex with NADP+, DAP, and R-naproxen | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Pedersen, L.C, London, R.E, Gabel, S.A, Krahn, J.M, DeRose, E.F. | | Deposit date: | 2019-12-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Structural Basis for Nonsteroidal Anti-Inflammatory Drug Inhibition of Human Dihydrofolate Reductase.
J.Med.Chem., 63, 2020
|
|
4UMC
 
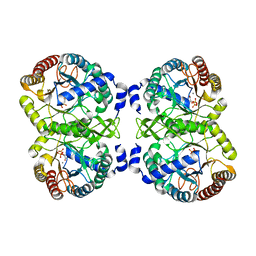 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | L-PHOSPHOLACTATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
6SMO
 
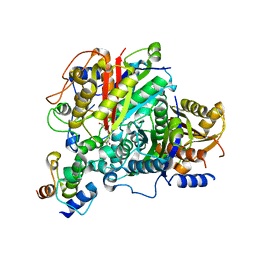 | | AntDE:AntF (apo): type II PKS acyl-carrier protein in complex with its ketosynthase bound to the hexaketide | | Descriptor: | 3,5,7,9,11-pentakis(oxidanylidene)dodecanal, Acyl carrier protein, Ketoacyl_synth_N domain-containing protein, ... | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
4UCG
 
 | | NmeDAH7PS R126S variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE, ... | | Authors: | Cross, P.J, Heyes, L.C, Zhang, S, Nazmi, A.R, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Functional Unit of Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase is Dimeric.
Plos One, 11, 2016
|
|
