4URH
 
 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4I85
 
 | | Crystal structure of transthyretin in complex with CHF5074 at neutral pH | | Descriptor: | 1-(3',4'-dichloro-2-fluorobiphenyl-4-yl)cyclopropanecarboxylic acid, Transthyretin | | Authors: | Zanotti, G, Cendron, L, Folli, C, Florio, P, Imbimbo, B.P, Berni, R. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors.
Febs Lett., 587, 2013
|
|
8IEJ
 
 | | RNF20-RNF40/hRad6A-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Ai, H, Deng, Z, Sun, M, Du, Y, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
1SXP
 
 | | BGT in complex with a 13mer DNA containing a central A:G mismatch | | Descriptor: | 5'-D(*A*AP*TP*AP*CP*TP*AP*AP*GP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*T)-3', DNA beta-glucosyltransferase, ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural evidence of a passive base flipping mechanism for {beta}-Glucosyltransferase
J.Biol.Chem., 279, 2004
|
|
4URE
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
8IEG
 
 | | Bre1(mRBD-RING)/Rad6-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1, Histone H2A type 1-B/E, ... | | Authors: | Ai, H, Deng, Z, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
1SVY
 
 | | SEVERIN DOMAIN 2, 1.75 ANGSTROM CRYSTAL STRUCTURE | | Descriptor: | CALCIUM ION, SEVERIN, SODIUM ION | | Authors: | Puius, Y.A, Fedorov, E.V, Eichinger, L, Sullivan, M, Schleicher, M, Almo, S.C. | | Deposit date: | 1998-08-10 | | Release date: | 1999-08-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the functional surface of domain 2 in the gelsolin superfamily.
Biochemistry, 39, 2000
|
|
8I0A
 
 | |
2IT7
 
 | |
6IRL
 
 | |
8IDO
 
 | | Crystal structure of nanobody VHH-T148 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T148, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
8IDM
 
 | | Crystal structure of nanobody VHH-227 with nanobody VHH-T71 and MERS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
8IDI
 
 | | Crystal structure of nanobody VHH-T71 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T71, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
8IEE
 
 | |
8IFN
 
 | | MERS-CoV spike trimer in complex with nanobody VHH-T148 | | Descriptor: | Spike glycoprotein, VHH-T148, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-19 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
8IS0
 
 | | Carbon Sulfoxide lyase - Y106F | | Descriptor: | 2-AMINO-ACRYLIC ACID, PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRY
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
2J5A
 
 | | Folding of S6 structures with divergent amino-acid composition: pathway flexibility within partly overlapping foldons | | Descriptor: | 30S RIBOSOMAL PROTEIN S6, SODIUM ION | | Authors: | Hansson, S, Olofsson, L, Hedberg, L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Folding of S6 Structures with Divergent Amino Acid Composition: Pathway Flexibility within Partly Overlapping Foldons.
J.Mol.Biol., 365, 2007
|
|
7B4R
 
 | | Structure of the 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus in complex with Coenzyme A and N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Maveyraud, L, Carivenc, C, Blanger, C, Mourey, L. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
7B4S
 
 | | Structure of the 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus in complex with Coenzyme A and compound 153786 from the NCI Open Database | | Descriptor: | 5-[(4-chlorophenyl)methyl]-6-[[2-(dimethylamino)ethylamino]methyl]-4-oxidanyl-1~{H}-pyrimidine-2-thione, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Maveyraud, L, Carivenc, C, Mourey, L. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
7NW3
 
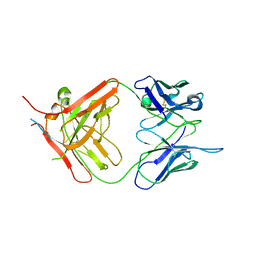 | | X-ray crystallographic study of PIYDIN, which contains the truncation determinants of binding PI and N, bound to RoAb13, a CCR5 antibody | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Saridakis, E, Helliwell, J.R, Govada, L, Chayen, N.E. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.200011 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
1JPS
 
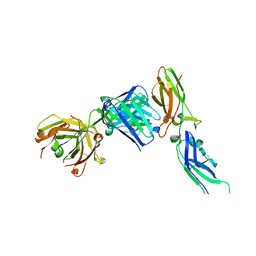 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
1JQ7
 
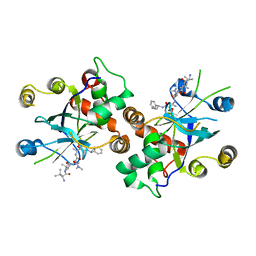 | | HCMV protease dimer-interface mutant, S225Y complexed to Inhibitor BILC 408 | | Descriptor: | ASSEMBLIN, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Batra, R, Khayat, R, Tong, L. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease.
Nat.Struct.Biol., 8, 2001
|
|
1JMC
 
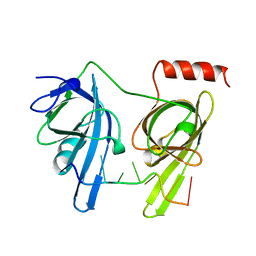 | | SINGLE STRANDED DNA-BINDING DOMAIN OF HUMAN REPLICATION PROTEIN A BOUND TO SINGLE STRANDED DNA, RPA70 SUBUNIT, RESIDUES 183-420 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), PROTEIN (REPLICATION PROTEIN A (RPA)) | | Authors: | Bochkarev, A, Pfuetzner, R, Edwards, A, Frappier, L. | | Deposit date: | 1996-11-11 | | Release date: | 1997-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA.
Nature, 385, 1997
|
|
