8OJS
 
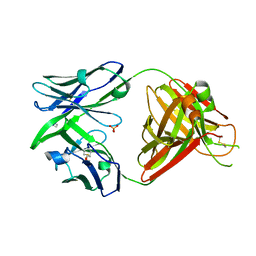 | | Crystal structure of the human IgD Fab - structure Fab1 | | Descriptor: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
3C49
 
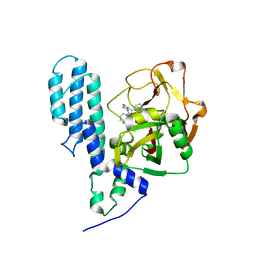 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor KU0058948 | | Descriptor: | 4-[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorobenzyl]phthalazin-1(2H)-one, Poly(ADP-ribose) polymerase 3 | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Helleday, T, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
3L3N
 
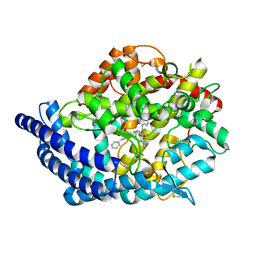 | | Testis ACE co-crystal structure with novel inhibitor lisW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Watermeyer, J.M, Kroger, W.L, O'Neil, H.G, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2009-12-17 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of domain-selective inhibitor binding in angiotensin-converting enzyme using a novel derivative of lisinopril.
Biochem.J., 428, 2010
|
|
3C4H
 
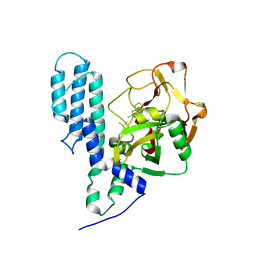 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor DR2313 | | Descriptor: | 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Poly(ADP-ribose) polymerase 3 | | Authors: | Lehtio, L, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
2F3J
 
 | |
4EGF
 
 | |
3THJ
 
 | |
3BZE
 
 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Hoare, H.L, Sullivan, L.C, Ely, L.K, Beddoe, T, Henderson, K.N, Lin, J, Clements, C.S, Reid, H.H, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-01-17 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors
J.Mol.Biol., 377, 2008
|
|
3C0D
 
 | | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1. Northeast Structural Genomics Consortium target VpR162 | | Descriptor: | Putative nitrite reductase NADPH (Small subunit) oxidoreductase protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Wang, D, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-19 | | Release date: | 2008-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1.
To be Published
|
|
3C3C
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate and L-CDP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
6OQS
 
 | | E. coli ATP synthase State 1b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7NLN
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with N-acetyl-glutamate | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
1LEK
 
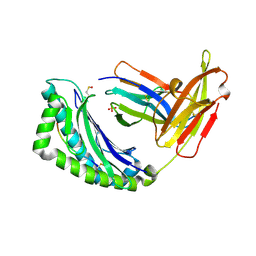 | | Crystal Structure of H-2Kbm3 bound to dEV8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Luz, J.G, Huang, M, Garcia, K.C, Rudolph, M.G, Apostolopoulos, V, Teyton, L, Wilson, I.A. | | Deposit date: | 2002-04-09 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(beta) Interactions.
J.Exp.Med., 195, 2002
|
|
6OQW
 
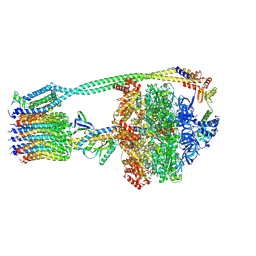 | | E. coli ATP synthase State 3a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OV6
 
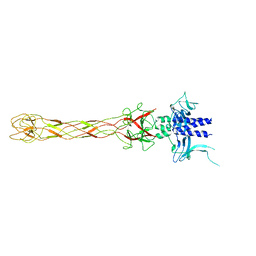 | | CRYSTALLOGRAPHIC STRUCTURE OF THE C24 PROTEIN FROM THE ANTARCTIC MICROORGANISM BIZIONIA ARGENTINENSIS | | Descriptor: | C24 PROTEIN, MANGANESE (II) ION | | Authors: | Klinke, S, Rinaldi, J, Guimaraes, B.G, Pellizza, L, Aran, M. | | Deposit date: | 2019-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the putative long tail fiber receptor-binding tip of a novel temperate bacteriophage from the Antarctic bacterium Bizionia argentinensis JUB59.
J.Struct.Biol., 212, 2020
|
|
3T5O
 
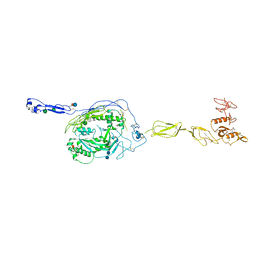 | | Crystal Structure of human Complement Component C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Complement component C6, ... | | Authors: | Aleshin, A.E, Stec, B, Bankston, L.A, DiScipio, R.G, Liddington, R.C. | | Deposit date: | 2011-07-27 | | Release date: | 2012-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.869 Å) | | Cite: | Structure of Complement C6 Suggests a Mechanism for Initiation and Unidirectional, Sequential Assembly of Membrane Attack Complex (MAC).
J.Biol.Chem., 287, 2012
|
|
4MS1
 
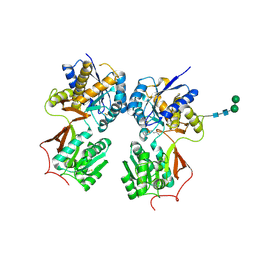 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP46381 | | Descriptor: | (S)-(3-aminopropyl)(cyclohexylmethyl)phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
6OQT
 
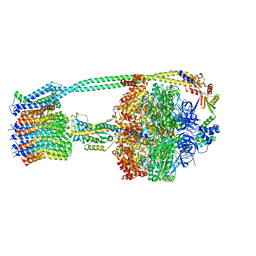 | | E. coli ATP synthase State 1c | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
5XB7
 
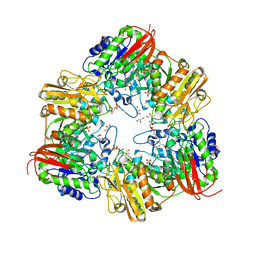 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
1LEG
 
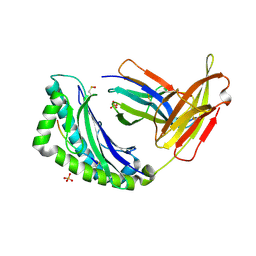 | | Crystal Structure of H-2Kb bound to the dEV8 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2-MICROGLOBULIN, ... | | Authors: | Luz, J.G, Huang, M, Garcia, K.C, Rudolph, M.G, Apostolopoulos, V, Teyton, L, Wilson, I.A. | | Deposit date: | 2002-04-09 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(beta) Interactions.
J.Exp.Med., 195, 2002
|
|
4EFS
 
 | | Human MMP12 in complex with L-glutamate motif inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Beau, F, Vera, L. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Simple pseudo-dipeptides with a P2' glutamate: a novel inhibitor family of matrix metalloproteases and other metzincins.
J.Biol.Chem., 287, 2012
|
|
6OWR
 
 | | NMR solution structure of YfiD | | Descriptor: | Autonomous glycyl radical cofactor | | Authors: | Bowman, S.E.J, Drennan, C.L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme.
J.Biol.Inorg.Chem., 24, 2019
|
|
3C5M
 
 | | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR199 | | Descriptor: | MANGANESE (II) ION, Oligogalacturonate lyase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Owens, L.A, Wang, D, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus.
To be Published
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
2QC8
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and methionine sulfoximine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Glutamine synthetase, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-19 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
