6OVC
 
 | | hMcl1 inhibitor complex | | Descriptor: | (2S)-N-(benzylsulfonyl)-4-(cyclobutylmethyl)-2-(2,4-dichlorophenyl)-3,4-dihydro-2H-1,4-benzoxazine-6-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Poppe, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6ZQI
 
 | | Cryo-EM structure of Spondweni virus prME | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6OQO
 
 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7MZL
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 210 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 210 heavy chain, PDI 210 light chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZG
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PDI 42 heavy chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZF
 
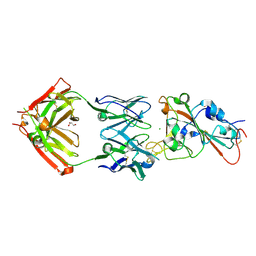 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 37 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZK
 
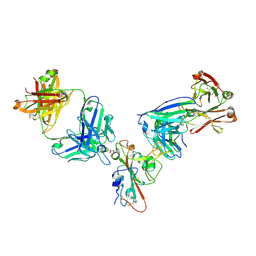 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 96 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZH
 
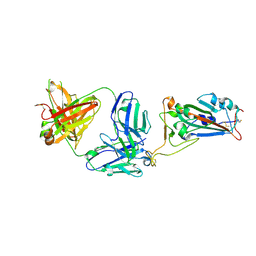 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 119 | | Descriptor: | Spike protein S1, WCSL 119 heavy chain, WCSL 119 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZN
 
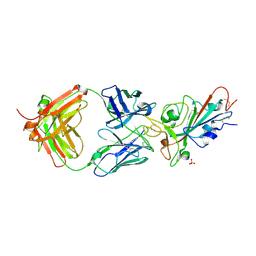 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 231 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 231 heavy chain, PDI 231 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZM
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 215 | | Descriptor: | ISOPROPYL ALCOHOL, PDI 215 heavy chain, PDI 215 light chain, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
1LED
 
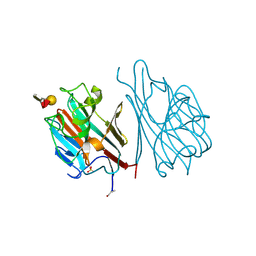 | | STRUCTURES OF THE LECTIN IV OF GRIFFONIA SIMPLICIFOLIA AND ITS COMPLEX WITH THE LEWIS B HUMAN BLOOD GROUP DETERMINANT AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Delbaere, L, Vandonselaar, M, Quail, J. | | Deposit date: | 1992-12-17 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis b human blood group determinant at 2.0 A resolution.
J.Mol.Biol., 230, 1993
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTT
 
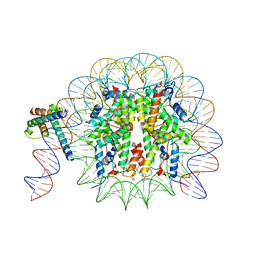 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | Descriptor: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSL
 
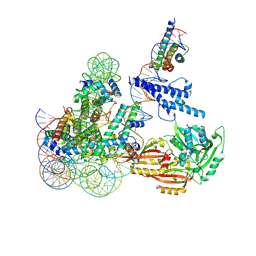 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTS
 
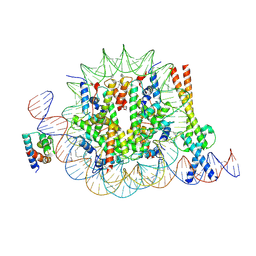 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
3BWN
 
 | | L-tryptophan aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
1UT1
 
 | | DraE adhesin from Escherichia Coli | | Descriptor: | 1,2-ETHANEDIOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
6OOG
 
 | |
1UXM
 
 | | A4V mutant of human SOD1 | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Hough, M.A, Grossmann, J.G, Antonyuk, S.V, Strange, R.W, Doucette, P.A, Rodriguez, J.A, Whitson, L.J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2004-02-26 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dimer Destabilization in Superoxide Dismutase May Result in Disease-Causing Properties: Structures of Motor Neuron Disease Mutants
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3BKL
 
 | | Testis ACE co-crystal structure with ketone ACE inhibitor kAW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Watermeyer, J.M, Kroger, W.L, O'Neill, H.G, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2007-12-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Probing the basis of domain-dependent inhibition using novel ketone inhibitors of Angiotensin-converting enzyme
Biochemistry, 47, 2008
|
|
4US4
 
 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
1UT2
 
 | | AfaE-3 adhesin from Escherichia Coli | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, SULFATE ION | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-02 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
6OUV
 
 | |
