7FIX
 
 | |
8H2U
 
 | | X-ray Structure of photosystem I-LHCI super complex from Chlamydomonas reinhardtii. | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, H, Kubota-Kawai, H, Misumi, Y, Kurisu, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
8HN3
 
 | | Soluble domain of cytochrome c-556 from Chlorobaculum tepidum | | Descriptor: | ACETATE ION, Cytochrome c-556, GLYCEROL, ... | | Authors: | Kishimoto, H, Azai, C, Yamamoto, T, Mutoh, R, Nakaniwa, T, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
8HN2
 
 | | Selenomethionine-labelled soluble domain of Rieske iron-sulfur protein from chlorobaculum tepidum | | Descriptor: | Cytochrome b6-f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Kishimoto, H, Mutoh, R, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
8I3J
 
 | | Crystal structure of human inner-arm dynein heavy chain d stalk and microtubule binding domain | | Descriptor: | Dynein axonemal heavy chain 1 | | Authors: | Ko, S, Yu, J, Toda, A, Tanaka, H, Kurisu, G. | | Deposit date: | 2023-01-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of the stalk region of axonemal inner-arm dynein-d reveals unique features in the coiled-coil and microtubule-binding domain.
Febs Lett., 597, 2023
|
|
1GEE
 
 | | Crystal structure of glucose dehydrogenase mutant Q252L complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-07 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
1G6K
 
 | | Crystal structure of glucose dehydrogenase mutant E96A complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
1VCL
 
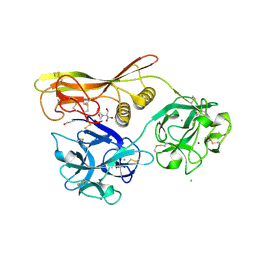 | | Crystal Structure of Hemolytic Lectin CEL-III | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
1IUE
 
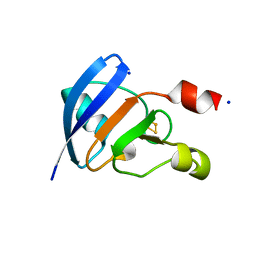 | |
1J18
 
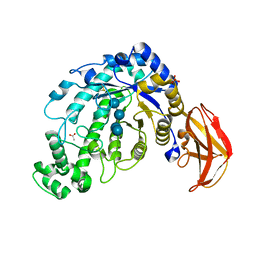 | | Crystal Structure of a Beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltose | | Descriptor: | ACETIC ACID, Beta-amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
1ITC
 
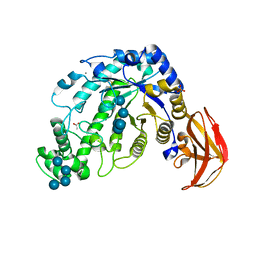 | | Beta-Amylase from Bacillus cereus var. mycoides Complexed with Maltopentaose | | Descriptor: | ACETIC ACID, Beta-Amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-01-17 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
5E37
 
 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | CALCIUM ION, EF-Hand domain-containing thioredoxin | | Authors: | Charoenwattansatien, R, Hochmal, A.K, Zinzius, K, Muto, R, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2015-10-02 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Calredoxin represents a novel type of calcium-dependent sensor-responder connected to redox regulation in the chloroplast
Nat Commun, 7, 2016
|
|
7CKA
 
 | |
5YGQ
 
 | |
5YXM
 
 | | Crystal structure of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Dynein light chain 1, axonemal, PHOSPHATE ION | | Authors: | Toda, A, Tanaka, H, Nishikawa, Y, Yagi, T, Kurisu, G. | | Deposit date: | 2017-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Structural atlas of dynein motors at atomic resolution.
Biophys Rev, 10, 2018
|
|
3VKH
 
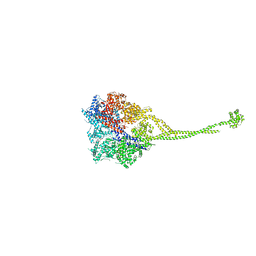 | | X-ray structure of a functional full-length dynein motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dynein heavy chain, cytoplasmic | | Authors: | Kon, T, Oyama, T, Shimo-Kon, R, Suto, K, Kurisu, G. | | Deposit date: | 2011-11-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The 2.8 A crystal structure of the dynein motor domain
Nature, 484, 2012
|
|
6AA2
 
 | | X-ray structure of ReQy1 (oxidized form) | | Descriptor: | Green fluorescent protein | | Authors: | Sugiura, K, Yasuda, A, Tabushi, N, Tanaka, H, Kurisu, G, Hisabori, T. | | Deposit date: | 2018-07-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multicolor redox sensor proteins can visualize redox changes in various compartments of the living cell.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
6A7K
 
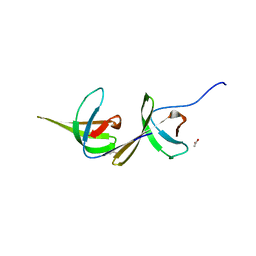 | | X-ray structure of NdhS from T. elongatus | | Descriptor: | ACETIC ACID, Tlr0636 protein | | Authors: | Umeno, K, Misumi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
8WIY
 
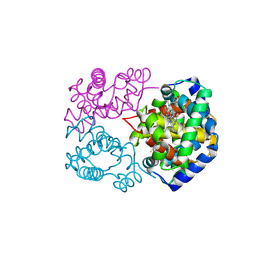 | | cryo-EM structure of alligator haemoglobin in oxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIZ
 
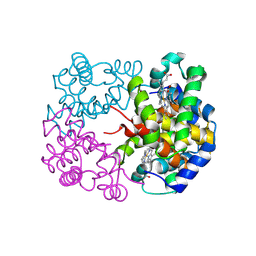 | | cryo-EM structure of alligator haemoglobin in deoxy form | | Descriptor: | BICARBONATE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ0
 
 | | cryo-EM structure of human haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ1
 
 | | cryo-EM structure of human haemoglobin in oxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIX
 
 | | cryo-EM structure of alligator haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ2
 
 | | cryo-EM structure of human haemoglobin in deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
5FJS
 
 | | Bacterial beta-glucosidase reveals the structural and functional basis of genetic defects in human glucocerebrosidase 2 (GBA2) | | Descriptor: | CALCIUM ION, GLUCOSYLCERAMIDASE | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Breen, I, Mutoha, R, Sansenya, S, Hua, Y, Tankrathok, A, Wu, L, Songsiriritthigul, C, Tanaka, H, Williams, S.J, Davies, G.J, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-10-12 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bacterial Beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (Gba2)
Acs Chem.Biol., 11, 2016
|
|
