1DQ7
 
 | | THREE-DIMENSIONAL STRUCTURE OF A NEUROTOXIN FROM RED SCORPION (BUTHUS TAMULUS) AT 2.2A RESOLUTION. | | Descriptor: | NEUROTOXIN | | Authors: | Sharma, M, Yadav, S, Karthikeyan, S, Kumar, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-12-30 | | Release date: | 2000-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional Structure of a Neurotoxin from Red Scorpion (Buthus tamulus) at 2.2A Resolution
To be Published
|
|
7BW9
 
 | |
8PQ4
 
 | |
8E1P
 
 | | Crystal structure of BG505 SOSIP.v4.1-GT1.2 trimer in complex with gl-PGV20 and PGT124 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505-SOSIP.v4.1-GT1.2gp120, ... | | Authors: | Sarkar, A, Kumar, S, Wilson, I.A. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.82 Å) | | Cite: | Germline-targeting HIV-1 Env vaccination induces VRC01-class antibodies with rare insertions.
Cell Rep Med, 4, 2023
|
|
8IYO
 
 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
8IYM
 
 | | Crystal structure of a protein acetyltransferase, HP0935 | | Descriptor: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
2SCP
 
 | |
1OYO
 
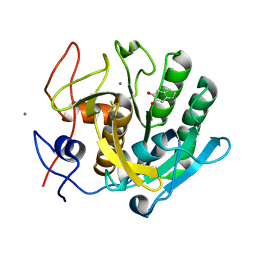 | | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution | | Descriptor: | 3H-INDOLE-5,6-DIOL, CALCIUM ION, Proteinase K | | Authors: | Singh, N, Sharma, S, Kumar, S, Raman, G, Singh, T.P. | | Deposit date: | 2003-04-06 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution
To be Published
|
|
3K1D
 
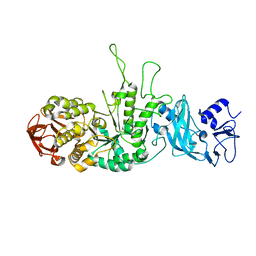 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
1QDF
 
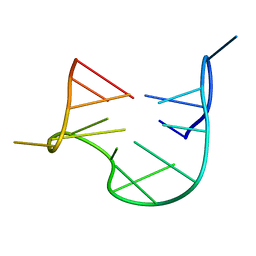 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, APTAMER (15MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDI
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, (12MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDH
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, APTAMER (15MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), MANGANESE (II) ION | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDK
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, (12MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), MANGANESE (II) ION | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
3OQT
 
 | | Crystal structure of Rv1498A protein from mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Rv1498A PROTEIN, SODIUM ION | | Authors: | Liu, F, Xiong, J, Kumar, S, Yang, C, Li, S, Ge, S, Xia, N, Swaminathan, K. | | Deposit date: | 2010-09-04 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A.
J.Struct.Biol., 175, 2011
|
|
8XEQ
 
 | |
8XGW
 
 | |
4HKE
 
 | | Crystal Structure of MoxT of Bacillus anthracis | | Descriptor: | Addiction module toxin component PemK, SULFATE ION | | Authors: | Verma, S, Kumar, S, Gourinath, S, Bhatnagar, R. | | Deposit date: | 2012-10-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of Bacillus anthracis MoxXT disruption and the modulation of MoxT ribonuclease activity by rationally designed peptides.
J.Biomol.Struct.Dyn., 33, 2015
|
|
8C3I
 
 | | Dark state of PAS-GAF fragment from Deinococcus radiodurans phytochrome | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Madan Kumar, S, Sebastian, W. | | Deposit date: | 2022-12-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dark state of PAS-GAF fragment from Deinococcus radiodurans phytochrome
To Be Published
|
|
4TUN
 
 | |
5VOE
 
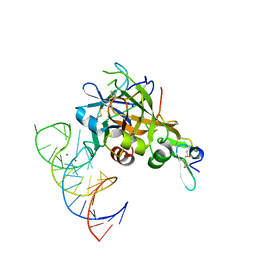 | | DesGla-XaS195A Bound to Aptamer 11F7t | | Descriptor: | Aptamer 11F7t (36-MER), CALCIUM ION, Coagulation factor X, ... | | Authors: | Gunaratne, R, Kumar, S, Frederiksen, J.W, Stayrook, S, Lohrmann, J.L, Perry, K, Chabata, C.V, Thalji, N.K, Ho, M.D, Arepally, G, Camire, R.M, Krishnaswamy, S.K, Sullenger, B.A. | | Deposit date: | 2017-05-02 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass.
Nat. Biotechnol., 36, 2018
|
|
5VOF
 
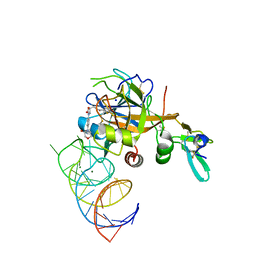 | | DesGla-XaS195A Bound to Aptamer 11F7t and Rivaroxaban | | Descriptor: | 5-chloro-N-({(5S)-2-oxo-3-[4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene-2-carboxamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Krishnaswamy, S, Kumar, S. | | Deposit date: | 2017-05-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass.
Nat. Biotechnol., 36, 2018
|
|
3PS7
 
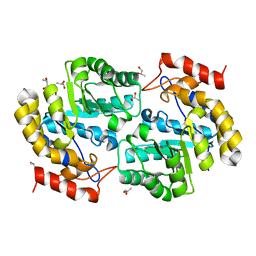 | | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Gautam, A, Kumar, S, Singh, A, Singh, N, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
3PH0
 
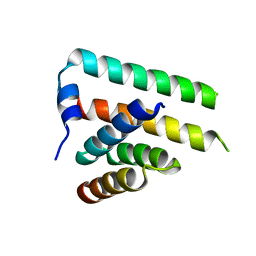 | | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila | | Descriptor: | AscE, AscG | | Authors: | Chatterjee, C, Kumar, S, Chakraborty, S, Tan, Y.W, Leung, K.Y, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila
Plos One, 6, 2011
|
|
8TQS
 
 | |
3B23
 
 | | Crystal structure of thrombin-variegin complex: Insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors | | Descriptor: | Thrombin heavy chain, Thrombin light chain, Variegin | | Authors: | Koh, C.Y, Kumar, S, Swaminathan, K, Kini, R.M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with s-variegin: insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors
Plos One, 6, 2011
|
|
