4QL8
 
 | | Crystal structure of Androgen Receptor in complex with the ligand | | Descriptor: | 2-chloro-4-[(3S,3aS,4S)-4-hydroxy-3-methoxy-3a,4,5,6-tetrahydro-3H-pyrrolo[1,2-b]pyrazol-2-yl]-3-methylbenzonitrile, Androgen receptor | | Authors: | Krishnamurthy, N, Sangeetha, R, Ghadiyaram, C, Sasmal, S, Subramanya, H.S. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3-alkoxy-pyrrolo[1,2-b]pyrazolines as selective androgen receptor modulators with ideal physicochemical properties for transdermal administration
J.Med.Chem., 57, 2014
|
|
1XFJ
 
 | | Crystal structure of protein CC_0490 from Caulobacter crescentus, Pfam DUF152 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-21 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a conserved hypothetical protein from Caulobacter crescentus
To be Published
|
|
1YBF
 
 | |
7C2W
 
 | |
7C2V
 
 | | Crystal Structure of IRAK4 kinase in complex with the inhibitor CA-4948 | | Descriptor: | 2-(2-methylpyridin-4-yl)-N-[2-morpholin-4-yl-5-[(3R)-3-oxidanylpyrrolidin-1-yl]-[1,3]oxazolo[4,5-b]pyridin-6-yl]-1,3-oxazole-4-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Krishnamurthy, N.R, Robert, B. | | Deposit date: | 2020-05-09 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of CA-4948, an Orally Bioavailable IRAK4 Inhibitor for Treatment of Hematologic Malignancies.
Acs Med.Chem.Lett., 11, 2020
|
|
1ZCC
 
 | | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens str.C58 | | Descriptor: | ACETATE ION, SULFATE ION, glycerophosphodiester phosphodiesterase | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-11 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens by SAD with a large asymmetric unit.
Proteins, 65, 2006
|
|
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
6INL
 
 | | Crystal structure of CDK2 IN complex with Inhibitor CVT-313 | | Descriptor: | 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), Cyclin-dependent kinase 2 | | Authors: | Talapati, S.R, Krishnamurthy, N.R. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of cyclin-dependent kinase 2 (CDK2) in complex with the specific and potent inhibitor CVT-313.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4JOA
 
 | | Crystal Structure of Human Anaplastic Lymphoma Kinase in complex with 7-azaindole based inhibitor | | Descriptor: | 3-[1-(2,5-difluorobenzyl)-1H-pyrazol-4-yl]-5-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, ALK tyrosine kinase receptor | | Authors: | Hosahalli, S, Krishnamurthy, N.R, Lakshminarasimhan, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-azaindole based anaplastic lymphoma kinase (ALK) inhibitors: wild type and mutant (L1196M) active compounds with unique binding mode
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5XMX
 
 | | Co-crystal structure of Inhibitor compound in complex with human PPARdelta LBD | | Descriptor: | (E)-6-[2-[[[4-(furan-2-yl)phenyl]carbonyl-methyl-amino]methyl]phenoxy]-4-methyl-hex-4-enoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Lakshminarasimhan, A, Rani, S.T, Senaiar, R.S, Krishnamurthy, N. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel highly selective peroxisome proliferator-activated receptor delta (PPAR delta) modulators with pharmacokinetic properties suitable for once-daily oral dosing.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5ZXI
 
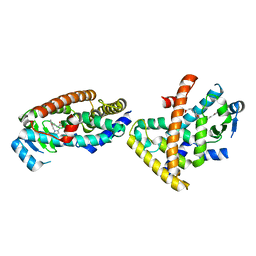 | | Co-crystal structure of an Inhibitor in complex with human PPARdelta LBD | | Descriptor: | 6-[2-({2-[4-(furan-2-yl)phenyl]-5-methyl-1H-imidazol-1-yl}methyl)phenoxy]hexanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Rani, S.T, Laxminarasimhan, A, Senaiar, R.S, Krishnamurthy, N. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective PPAR delta Modulators Improve Mitochondrial Function: Potential Treatment for Duchenne Muscular Dystrophy (DMD).
ACS Med Chem Lett, 9, 2018
|
|
7F80
 
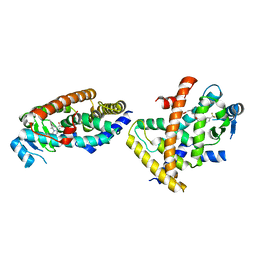 | |
6AH9
 
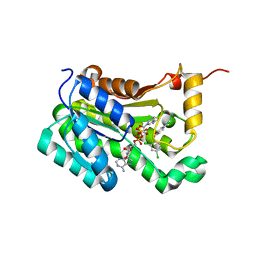 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and Triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and Triclosan
To Be Published
|
|
1U0F
 
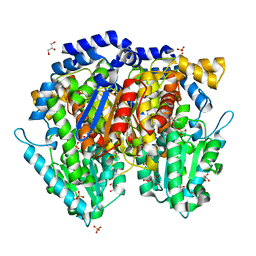 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0G
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0E
 
 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
6AHE
 
 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ternary complex formation of AFN-1252 with Acinetobacter baumannii FabI and NADH: Crystallographic and biochemical studies.
Chem.Biol.Drug Des., 96, 2020
|
|
