2YNM
 
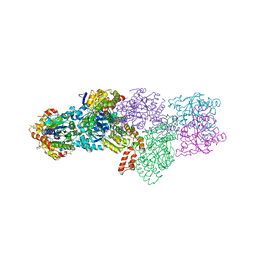 | | Structure of the ADPxAlF3-Stabilized Transition State of the Nitrogenase-like Dark-Operative Protochlorophyllide Oxidoreductase Complex from Prochlorococcus marinus with Its Substrate Protochlorophyllide a | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Krausze, J, Lange, C, Heinz, D.W, Moser, J. | | Deposit date: | 2012-10-16 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Adp-Aluminium Fluoride-Stabilized Protochlorophyllide Oxidoreductase Complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8RZ6
 
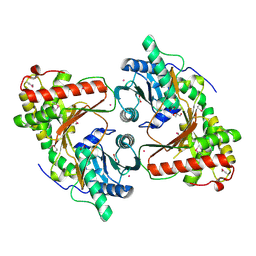 | | SeMet derivative structure of the condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | FORMIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
8QNF
 
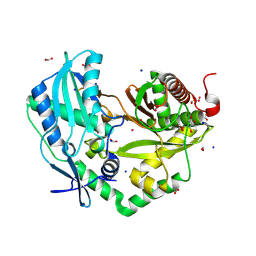 | | Crystal structure of the Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase, FORMIC ACID, GLYCEROL, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
4UZB
 
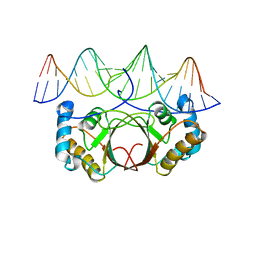 | | KSHV LANA (ORF73) C-terminal domain mutant bound to LBS1 DNA (R1039Q, R1040Q, K1055E, K1109A, D1110A, A1121E, K1138S, K1140D, K1141D) | | Descriptor: | LANA BINDING SITE 1 DNA, ORF 73 | | Authors: | Hellert, J, Krausze, J, Luhrs, T. | | Deposit date: | 2014-09-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.865 Å) | | Cite: | The 3D Structure of Kaposi Sarcoma Herpesvirus Lana C-Terminal Domain Bound to DNA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UZC
 
 | |
4A4V
 
 | | Ligand binding domain of human PPAR gamma in complex with amorfrutin 2 | | Descriptor: | AMORFRUTIN 2, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | de Groot, J.C, Weidner, C, Krausze, J, Kawamoto, K, Schroeder, F.C, Sauer, S, Buessow, K. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Amorfrutins Bound to the Peroxisome Proliferator-Activated Receptor Gamma.
J.Med.Chem., 56, 2013
|
|
4A4W
 
 | | Ligand binding domain of human PPAR gamma in complex with amorfrutin B | | Descriptor: | AMORFRUTIN B, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | de Groot, J.C, Weidner, C, Krausze, J, Kawamoto, K, Schroeder, F.C, Sauer, S, Buessow, K. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Amorfrutins Bound to the Peroxisome Proliferator-Activated Receptor Gamma.
J.Med.Chem., 56, 2013
|
|
4D3Z
 
 | |
4D3X
 
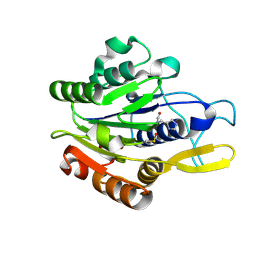 | |
4D3Y
 
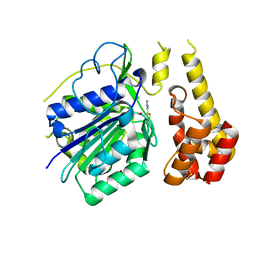 | |
2YBO
 
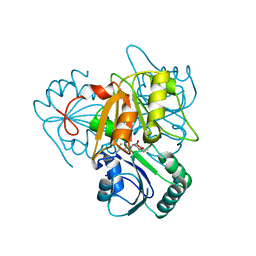 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-08 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
2YBQ
 
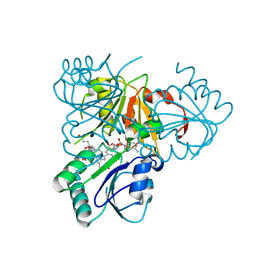 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
2YQ0
 
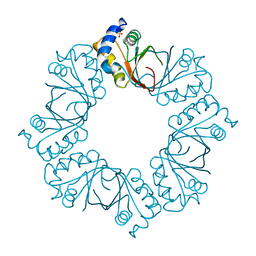 | |
2YQ1
 
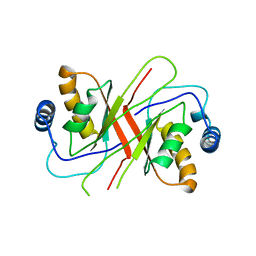 | |
2YPZ
 
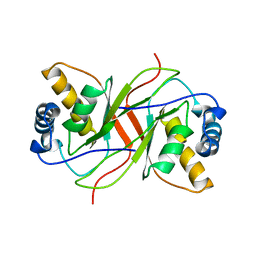 | |
2YPY
 
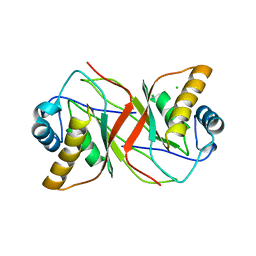 | |
