6WUS
 
 | |
6WUR
 
 | |
1G9L
 
 | | SOLUTION STRUCTURE OF THE PABC DOMAIN OF HUMAN POLY(A) BINDING PROTEIN | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1 | | Authors: | Kozlov, G, Trempe, J.-F, Khaleghpour, K, Kahvejian, A, Ekiel, I, Gehring, K. | | Deposit date: | 2000-11-24 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the C-terminal PABC domain of human poly(A)-binding protein.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GH9
 
 | |
3BCY
 
 | | Crystal structure of YER067W | | Descriptor: | Protein YER067W | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional study of YER067W, a new protein involved in yeast metabolism control and drug resistance.
Plos One, 5, 2010
|
|
5BTY
 
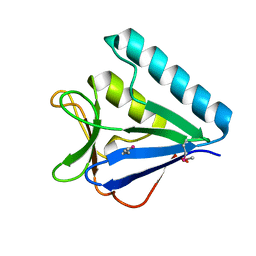 | | Structure of the middle domain of lpg1496 from Legionella pneumophila in P21 space group | | Descriptor: | lpg1496 | | Authors: | Kozlov, G, Zhang, Y, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
1RRZ
 
 | |
8EY6
 
 | |
4EEW
 
 | |
1D5G
 
 | |
4G3O
 
 | |
3ICI
 
 | |
4I6X
 
 | |
3BXY
 
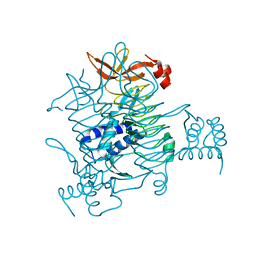 | | Crystal structure of tetrahydrodipicolinate N-succinyltransferase from E. coli | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli tetrahydrodipicolinate N-succinyltransferase reveals the role of a conserved C-terminal helix in cooperative substrate binding.
Febs Lett., 582, 2008
|
|
5KDG
 
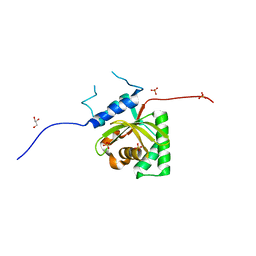 | | Crystal Structure of Salmonella Typhimurium Effector GtgE | | Descriptor: | GLYCEROL, Gifsy-2 prophage protein, SULFATE ION | | Authors: | Kozlov, G, Xu, C, Wong, K, Gehring, K, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the Salmonella Typhimurium Effector GtgE.
PLoS ONE, 11, 2016
|
|
8EY7
 
 | |
8EY8
 
 | |
8G91
 
 | |
8G90
 
 | |
3GZH
 
 | |
6B3Y
 
 | | Crystal structure of the PH-like domain from DENND3 | | Descriptor: | DENN domain-containing protein 3 | | Authors: | Kozlov, G, Xu, J, Menade, M, Beaugrand, M, Pan, T, McPherson, P.S, Gehring, K. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | A PH-like domain of the Rab12 guanine nucleotide exchange factor DENND3 binds actin and is required for autophagy.
J. Biol. Chem., 293, 2018
|
|
3IDV
 
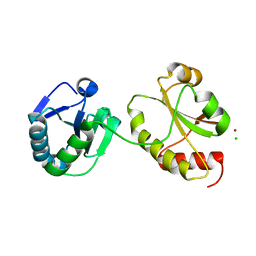 | | Crystal structure of the a0a fragment of ERp72 | | Descriptor: | CHLORIDE ION, Protein disulfide-isomerase A4, ZINC ION | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2009-07-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Catalytic a(0)a Fragment of the Protein Disulfide Isomerase ERp72.
J.Mol.Biol., 401, 2010
|
|
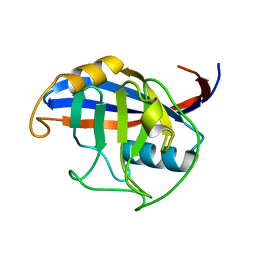
3ICH
 
 | |
3PDZ
 
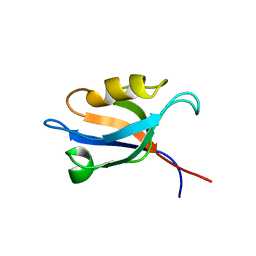 | |
3RG0
 
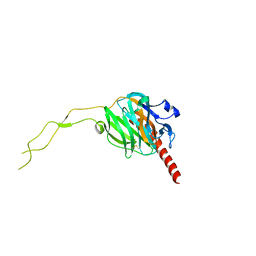 | | Structural and functional relationships between the lectin and arm domains of calreticulin | | Descriptor: | CALCIUM ION, Calreticulin | | Authors: | Kozlov, G, Pocanschi, C.L, Brockmeier, U, Williams, D.B, Gehring, K. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and Functional Relationships between the Lectin and Arm Domains of Calreticulin.
J.Biol.Chem., 286, 2011
|
|
