4NL2
 
 | |
4NL3
 
 | |
4KM7
 
 | | Human folate receptor alpha (FOLR1) at acidic pH, triclinic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Folate receptor alpha, POTASSIUM ION | | Authors: | Kovach, A.R, Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2X5O
 
 | | Discovery of Novel 5-Benzylidenerhodanine- and 5-Benzylidene- thiazolidine-2,4-dione Inhibitors of MurD Ligase | | Descriptor: | AZIDE ION, CHLORIDE ION, N-({3-[({4-[(Z)-(2,4-DIOXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL}AMINO)METHYL]PHENYL}CARBONYL)-D-GLUTAMIC ACID, ... | | Authors: | Zidar, N, Tomasic, T, Sink, R, Rupnik, V, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelja, D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of novel 5-benzylidenerhodanine and 5-benzylidenethiazolidine-2,4-dione inhibitors of MurD ligase.
J. Med. Chem., 53, 2010
|
|
2WJP
 
 | | CRYSTAL STRUCTURE OF MURD LIGASE IN COMPLEX WITH D-GLU CONTAINING RHODANINE INHIBITOR | | Descriptor: | AZIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Rupnik, V, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Muller-Premru, M, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2009-05-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel 5-Benzylidenerhodanine and 5-Benzylidenethiazolidine-2,4-Dione Inhibitors of Murd Ligase.
J.Med.Chem., 53, 2010
|
|
2Y68
 
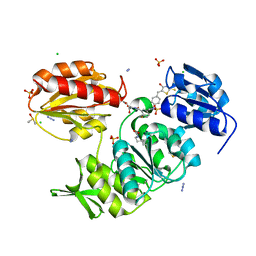 | | Structure-based design of a new series of D-glutamic acid-based inhibitors of bacterial MurD ligase | | Descriptor: | 2-[[2-fluoro-5-[[[4-[(Z)-(4-oxo-2-sulfanylidene-1,3-thiazolidin-5-ylidene)methyl]phenyl]amino]methyl]phenyl]carbonylamino]pentanedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-based design of a new series of D-glutamic acid based inhibitors of bacterial UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (MurD).
J. Med. Chem., 54, 2011
|
|
2Y66
 
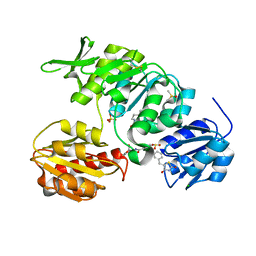 | | New 5-Benzylidenethiazolidine-4-one Inhibitors of Bacterial MurD Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation | | Descriptor: | (2R)-2-[[3-[[3-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]phenoxy]methyl]phenyl]carbonylamino]pentanedioic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Zidar, N, Tomasic, T, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | New 5-Benzylidenethiazolidin-4-One Inhibitors of Bacterial Murd Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation.
Eur.J.Med.Chem, 46, 2011
|
|
2Y67
 
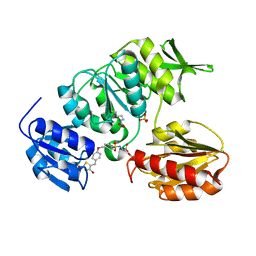 | | New 5-Benzylidenethiazolidine-4-one Inhibitors of Bacterial MurD Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation | | Descriptor: | (2R)-2-[[4-[[4-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]phenoxy]methyl]phenyl]sulfonylamino]pentanedioic acid, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Zidar, N, Tomasic, T, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 5-Benzylidenethiazolidin-4-One Inhibitors of Bacterial Murd Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation.
Eur.J.Med.Chem, 46, 2011
|
|
2Y1O
 
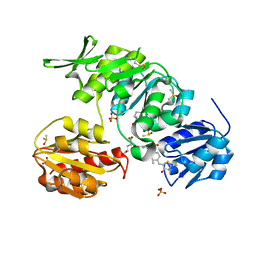 | | Dual-target Inhibitor of MurD and MurE Ligases: Design, Synthesis and Binding Mode Studies | | Descriptor: | (2R)-2-[[3-[[4-[(Z)-(4-OXO-2-SULFANYLIDENE-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL]METHYLAMINO]PHENYL]CARBONYLAMINO]PENTANEDIOIC ACID, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Tomasic, T, Sink, R, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Kikelj, D, Peterlin-Masic, L. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Dual Inhibitor of MurD and MurE Ligases from Escherichia coli and Staphylococcus aureus.
ACS Med Chem Lett, 3, 2012
|
|
4B2O
 
 | | Crystal structure of Bacillus subtilis YmdB, a global regulator of late adaptive responses. | | Descriptor: | FE (II) ION, PHOSPHATE ION, YMDB PHOSPHODIESTERASE | | Authors: | Newman, J.A, Diethmaier, C, Kovacs, A.T, Rodrigues, C, Kuipers, O.P, Stulke, J, Lewis, R.J. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Ymdb Phosphodiesterase is a Global Regulator of Late Adaptive Responses in Bacillus Subtilis.
J.Bacteriol., 196, 2014
|
|
1QGE
 
 | | NEW CRYSTAL FORM OF PSEUDOMONAS GLUMAE (FORMERLY CHROMOBACTERIUM VISCOSUM ATCC 6918) LIPASE | | Descriptor: | CALCIUM ION, PROTEIN (TRIACYLGLYCEROL HYDROLASE) | | Authors: | Lang, D.A, Stadler, P, Kovacs, A, Paltauf, F, Dijkstra, B.W. | | Deposit date: | 1999-04-27 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Kinetic Investigations of Enantiomeric Binding Mode of Subclass I Lipases from the Family of Pseudomonadaceae
To be Published
|
|
4ESF
 
 | |
4ESB
 
 | | Crystal structure of PadR-like transcriptional regulator (BC4206) from Bacillus cereus strain ATCC 14579 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Fibriansah, G, Kovacs, A.T, Kuipers, O.P, Thunnissen, A.M.W.H. | | Deposit date: | 2012-04-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Two Transcriptional Regulators from Bacillus cereus Define the Conserved Structural Features of a PadR Subfamily.
Plos One, 7, 2012
|
|
3NMH
 
 | | Crystal structure of the abscisic receptor PYL2 in complex with pyrabactin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6BS3
 
 | | Crystal structure of ADP-bound bacterial Get3-like A and B in Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anion transporter, CALCIUM ION, ... | | Authors: | Li, H, Hu, K, Kovach, A. | | Deposit date: | 2017-12-01 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Guided Entry of Tail-Anchored Proteins 3 Homologues in Mycobacterium tuberculosis.
J.Bacteriol., 201, 2019
|
|
6BS5
 
 | | Crystal structure of AMP-PNP-bound bacterial Get3-like A and B in Mycobacterium tuberculosis | | Descriptor: | Anion transporter, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, H, Hu, K, Kovach, A. | | Deposit date: | 2017-12-01 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterization of Guided Entry of Tail-Anchored Proteins 3 Homologues in Mycobacterium tuberculosis.
J.Bacteriol., 201, 2019
|
|
6BS4
 
 | | Crystal structure of ATPgammaS-bound bacterial Get3-like A and B in Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anion transporter, MAGNESIUM ION, ... | | Authors: | Li, H, Hu, K, Kovach, A. | | Deposit date: | 2017-12-01 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of Guided Entry of Tail-Anchored Proteins 3 Homologues in Mycobacterium tuberculosis.
J.Bacteriol., 201, 2019
|
|
7LHG
 
 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapG in the first conformation | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, P fimbria tip G-adhesin PapG-II, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
7LHH
 
 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapG in the second conformation | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, P fimbria tip G-adhesin PapG-II, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
7LHI
 
 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapF-PapG | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, Fimbrial protein, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
7KY7
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the apo E1 state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KYA
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E2P state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KY8
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E1-ATP state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KYC
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf1-Lem3 complex in the E2P state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KY5
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E2P transition state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
