8EAC
 
 | | Thermus thermophilus methylenetetrahydrofolate reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase | | Authors: | Yamada, K, Koutmos, M. | | Deposit date: | 2022-08-28 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 5-Formyltetrahydrofolate promotes conformational remodeling in a methylenetetrahydrofolate reductase active site and inhibits its activity.
J.Biol.Chem., 299, 2022
|
|
6WGV
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to adenosylcobalamin and PPPi | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7TH4
 
 | |
7TH5
 
 | | Thermus thermophilus methylenetetrahydrofolate reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase, SODIUM ION | | Authors: | Yamada, K, Koutmos, M. | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 5-Formyltetrahydrofolate promotes conformational remodeling in a methylenetetrahydrofolate reductase active site and inhibits its activity.
J.Biol.Chem., 299, 2022
|
|
3IV9
 
 | |
7RUU
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUT
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Corrinoid adenosyltransferase, GLYCEROL, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUV
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase E193K bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
3IVA
 
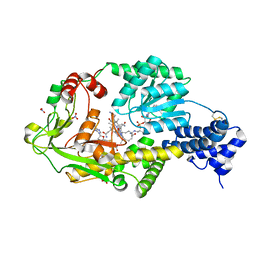 | | Structure of the B12-dependent Methionine Synthase (MetH) C-teminal half with AdoHcy bound | | Descriptor: | COBALAMIN, Methionine synthase, NITRATE ION, ... | | Authors: | Pattridge, K.A, Koutmos, M, Smith, J.L. | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the reactivation of cobalamin-dependent methionine synthase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5DIZ
 
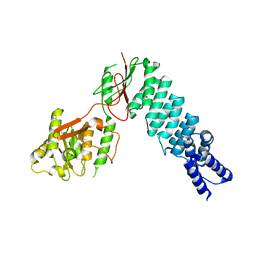 | | Crystal Structure of nuclear proteinaceous RNase P 2 (PRORP2) from A. thaliana | | Descriptor: | Proteinaceous RNase P 2, ZINC ION | | Authors: | Karasik, A, Shanmuganathan, A, Howard, M.J, Fierke, C.A, Koutmos, M. | | Deposit date: | 2015-09-01 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nuclear Protein-Only Ribonuclease P2 Structure and Biochemical Characterization Provide Insight into the Conserved Properties of tRNA 5' End Processing Enzymes.
J.Mol.Biol., 428, 2016
|
|
8SSG
 
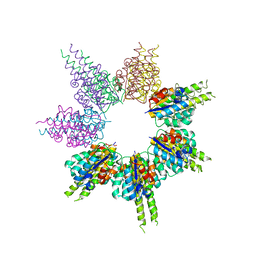 | |
8SSE
 
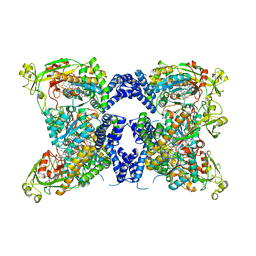 | | Methionine synthase, C-terminal fragment, Cobalamin and Reactivation Domains from Thermus thermophilus HB8 | | Descriptor: | COBALAMIN, Methionine synthase | | Authors: | Yamada, K, Mendoza, J, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of full-length cobalamin-dependent methionine synthase and cofactor loading captured in crystallo.
Nat Commun, 14, 2023
|
|
6WH5
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to cob(II)alamin and PPPi | | Descriptor: | COBALAMIN, Corrinoid adenosyltransferase, MAGNESIUM ION, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-07 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5CUZ
 
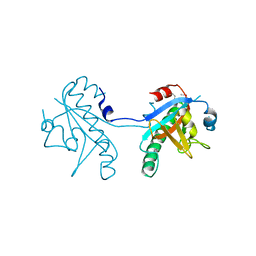 | | Crystal structure of SeMet-substituted N-terminal truncated human B12-chaperone CblD (108-296) | | Descriptor: | Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Yamada, K, Gherasim, C, Banerjee, R, Koutmos, M. | | Deposit date: | 2015-07-25 | | Release date: | 2015-09-23 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Human B12 Trafficking Protein CblD Reveals Molecular Mimicry and Identifies a New Subfamily of Nitro-FMN Reductases.
J.Biol.Chem., 290, 2015
|
|
5CV0
 
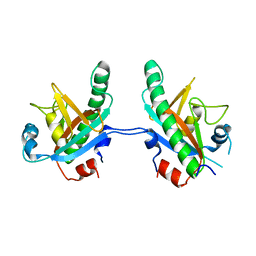 | | Crystal structure of N-terminal truncated human B12-chaperone CblD (108-296) | | Descriptor: | Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Yamada, K, Gherasim, C, Banerjee, R, Koutmos, M. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human B12 Trafficking Protein CblD Reveals Molecular Mimicry and Identifies a New Subfamily of Nitro-FMN Reductases.
J.Biol.Chem., 290, 2015
|
|
6WGU
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase | | Descriptor: | Corrinoid adenosyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8SSF
 
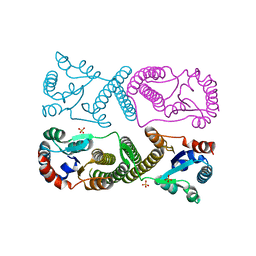 | | Minimal protein-only/RNA-free Ribonuclease P from Hydrogenobacter thermophilus | | Descriptor: | RNA-free ribonuclease P, SULFATE ION | | Authors: | Mendoza, J, Mallik, L, Wilhelm, C.A, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial RNA-free RNase P: Structural and functional characterization of multiple oligomeric forms of a minimal protein-only ribonuclease P.
J.Biol.Chem., 299, 2023
|
|
8SSC
 
 | |
8SSD
 
 | |
6XJJ
 
 | | Structure of non-heme iron enzyme TropC: Radical tropolone biosynthesis | | Descriptor: | 2-oxoglutarate-dependent dioxygenase tropC, ACETATE ION, FE (III) ION, ... | | Authors: | Mallik, L, Doyon, T.J, Narayan, A.R.H, Koutmos, M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Radical Tropolone Biosynthesis
Chemrxiv, 2020
|
|
6WGS
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8UY2
 
 | | Methylenetetrahydrofolate reductase from Chaetomium thermophilum DSM 1495, AdoMet-bound, Inhibited (T) State | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yamada, K, Mendoza, J, Koutmos, M. | | Deposit date: | 2023-11-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural basis of S-adenosylmethionine-dependent allosteric transition from active to inactive states in methylenetetrahydrofolate reductase.
Nat Commun, 15, 2024
|
|
5UOS
 
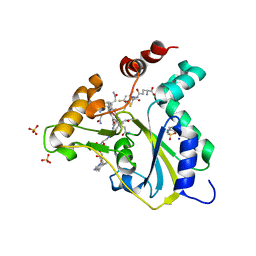 | | Crystal Structure of CblC (MMACHC) (1-238), a human B12 processing enzyme, complexed with an Antivitamin B12 | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, 2-PHENYLAMINO-ETHANESULFONIC ACID, COBALAMIN, ... | | Authors: | Shanmuganathan, A, Karasik, A, Ruetz, M, Banerjee, R, Krautler, B, Koutmos, M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Antivitamin B12 Inhibition of the Human B12 -Processing Enzyme CblC: Crystal Structure of an Inactive Ternary Complex with Glutathione as the Cosubstrate.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6X8Z
 
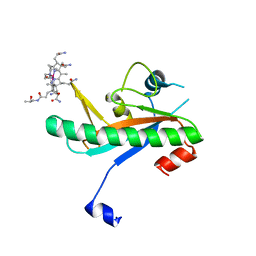 | | Crystal structure of N-truncated human B12 chaperone CblD(C262S)-thiolato-cob(III)alamin complex (108-296) | | Descriptor: | COBALAMIN, Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Mascarenhas, R, Li, Z, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interprotein Co-S Coordination Complex in the B 12 -Trafficking Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6BV5
 
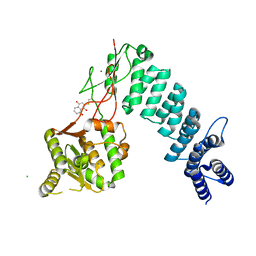 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana after 45-minute soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
