8R7Q
 
 | |
8R7P
 
 | |
6R4O
 
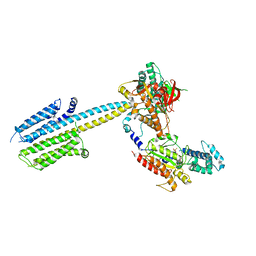 | | Structure of a truncated adenylyl cyclase bound to MANT-GTP, forskolin and an activated stimulatory Galphas protein | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase 9, ... | | Authors: | Qi, C, Sorrentino, S, Medalia, O, Korkhov, V.M. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein.
Science, 364, 2019
|
|
7ZXO
 
 | |
7ZXQ
 
 | |
7ZXP
 
 | |
7ZXM
 
 | |
7ZXT
 
 | |
7PDH
 
 | |
7PD4
 
 | |
7PD8
 
 | |
7PDE
 
 | |
7PDD
 
 | |
7PDG
 
 | |
7PDF
 
 | |
7ZXN
 
 | | cryo-EM structure of Connexin 32 gap junction channel | | Descriptor: | CHOLESTEROL, Gap junction beta-1 protein | | Authors: | Qi, C, Korkhov, V.M. | | Deposit date: | 2022-05-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structures of wild-type and selected CMT1X mutant connexin 32 gap junction channels and hemichannels.
Sci Adv, 9, 2023
|
|
5M34
 
 | | Structure of cobinamide-bound BtuF mutant W66Y, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
5M3B
 
 | | Structure of cobinamide-bound BtuF mutant W66L, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
5M29
 
 | | Structure of cobinamide-bound BtuF, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
