4WZP
 
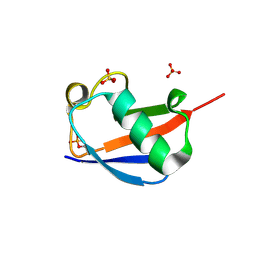 | | Ser65 phosphorylated ubiquitin, major conformation | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Wauer, T, Wagstaff, J, Freund, S.M.V, Komander, D. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis.
Embo J., 34, 2015
|
|
3F89
 
 | | NEMO CoZi domain | | Descriptor: | NF-kappa-B essential modulator | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
6SAK
 
 | |
5C9V
 
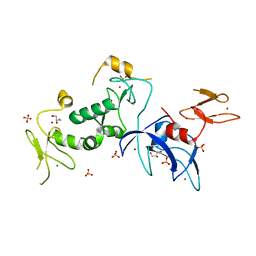 | | Structure of human Parkin G319A | | Descriptor: | E3 ubiquitin-protein ligase parkin, GLYCEROL, SULFATE ION, ... | | Authors: | Wauer, T, Komander, D. | | Deposit date: | 2015-06-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of phospho-ubiquitin-induced PARKIN activation.
Nature, 524, 2015
|
|
5OXI
 
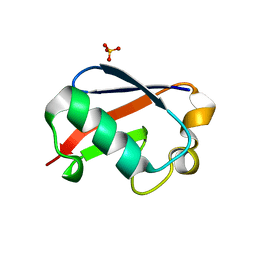 | | C-terminally retracted ubiquitin L67S mutant | | Descriptor: | SULFATE ION, Ubiquitin L67S mutant | | Authors: | Gladkova, C.G, Schubert, A.F, Wagstaff, J.L, Pruneda, J.N, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
5OXH
 
 | | C-terminally retracted ubiquitin T66V/L67N mutant | | Descriptor: | SULFATE ION, Ubiquitin T66V/L67N mutant | | Authors: | Gladkova, C, Schubert, A.F, Wagstaff, J.L, Pruneda, J.P, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
5OHM
 
 | | K33-specific affimer bound to K33 diUb | | Descriptor: | K33-specific affimer, POLYETHYLENE GLYCOL (N=34), Polyubiquitin-C | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
7OWD
 
 | |
7OWC
 
 | |
9B12
 
 | |
9AVT
 
 | | Structure of TAB2 NZF domain bound to K6 / Lys6-linked diubiquitin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TGF-beta-activated kinase 1 and MAP3K7-binding protein 2, ... | | Authors: | Michel, M.A, Scutts, S, Komander, D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of TAB2 NZF domain bound to K6 / Lys6-linked diubiquitin
To be published
|
|
9AVW
 
 | |
6XAA
 
 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
9B0B
 
 | |
9B0Z
 
 | |
9AZJ
 
 | |
8UYF
 
 | | Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | Serine/threonine-protein kinase Pink1, mitochondrial | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
8UYI
 
 | | Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase Pink1, ... | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
4OYK
 
 | |
5CAW
 
 | |
8UYH
 
 | | Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Pink1, ... | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
6W9O
 
 | |
4OYJ
 
 | | Structure of the apo HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, SULFATE ION | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2014-02-12 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Basis and Regulation of OTULIN-LUBAC Interaction.
Mol.Cell, 54, 2014
|
|
6W9S
 
 | |
