1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
1ISE
 
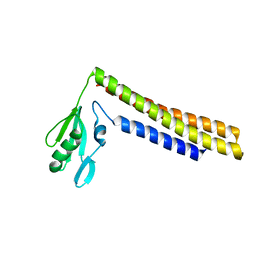 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
1IY5
 
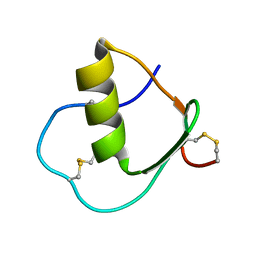 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1IY6
 
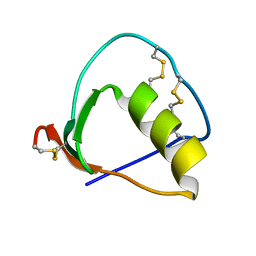 | | Solution structure of OMSVP3 variant, P14C/N39C | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
6LOS
 
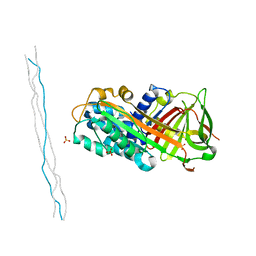 | | Crystal structure of mouse PEDF in complex with heterotrimeric collagen model peptide. | | Descriptor: | Collagen model peptide, type I, alpha 1, ... | | Authors: | Kawahara, K, Maruno, T, Oki, H, Yoshida, T, Ohkubo, T, Koide, T, Kobayashi, Y. | | Deposit date: | 2020-01-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Spatiotemporal regulation of PEDF signaling by type I collagen remodeling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1IW4
 
 | | Solution structure of ascidian trypsin inhibitor | | Descriptor: | trypsin inhibitor | | Authors: | Hemmi, H, Yoshida, T, Kumazaki, T, Nemoto, N, Hasegawa, J, Nishioka, F, Kyogoku, Y, Yokosawa, H, Kobayashi, Y. | | Deposit date: | 2002-04-19 | | Release date: | 2002-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ascidian trypsin inhibitor determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 41, 2002
|
|
3AHA
 
 | | Crystal structure of the complex between gp41 fragments N36 and C34 mutant N126K/E137Q | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Transmembrane protein gp41 | | Authors: | Izumi, K, Nakamura, S, Nakano, H, Shimura, K, Sakagami, Y, Oishi, S, Uchiyama, S, Ohkubo, T, Kobayashi, Y, Fujii, N, Matsuoka, M, Kodama, E.N. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino terminal heptad repeat.
Antiviral Res., 2010
|
|
2WOQ
 
 | | Porphobilinogen Synthase (HemB) in Complex with 5-acetamido-4- oxohexanoic acid (Alaremycin 2) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALAREMYCIN 2, ... | | Authors: | Heinemann, I.U, Schulz, C, Schubert, W.-D, Heinz, D.W, Wang, Y.-G, Kobayashi, Y, Awa, Y, Wachi, M, Jahn, D, Jahn, M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the heme biosynthetic Pseudomonas aeruginosa porphobilinogen synthase in complex with the antibiotic alaremycin.
Antimicrob. Agents Chemother., 54, 2010
|
|
3AQB
 
 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium | | Descriptor: | CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, Component B of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
3AQC
 
 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium and FPP analogue | | Descriptor: | (2E,6E)-7,11-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2Z6W
 
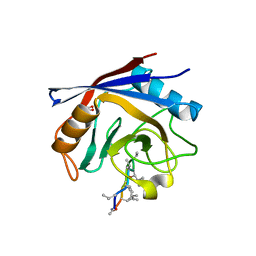 | | Crystal structure of human cyclophilin D in complex with cyclosporin A | | Descriptor: | CITRIC ACID, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Kajitani, K, Fujihashi, M, Kobayashi, Y, Shimizu, S, Tsujimoto, Y, Miki, K. | | Deposit date: | 2007-08-09 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal Structure of Human Cyclophilin D in Complex with its Inhibitor, Cyclosporin a at 0.96-A Resolution.
Proteins, 70, 2008
|
|
2Z2T
 
 | |
2PA2
 
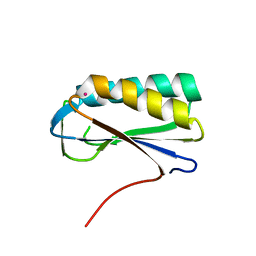 | | Crystal structure of human Ribosomal protein L10 core domain | | Descriptor: | 60S ribosomal protein L10, POTASSIUM ION | | Authors: | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
3WJ4
 
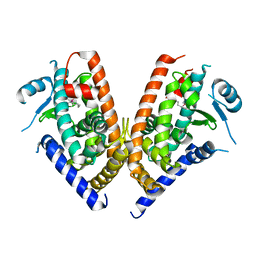 | | Crystal structure of PPARgamma ligand binding domain in complex with tributyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, tributylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3VOR
 
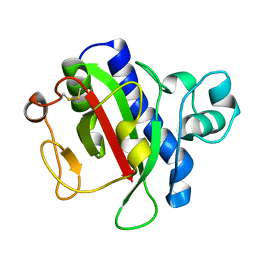 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2AI5
 
 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-07-29 | | Release date: | 2006-05-23 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
2D0S
 
 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
1VA1
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1WZB
 
 | | Crystal structure of the collagen triple helix model [{HYP(R)-HYP(R)-GLY}10]3 | | Descriptor: | Collagen triple helix | | Authors: | Kawahara, K, Nakamura, S, Nishi, Y, Uchiyama, S, Nishiuchi, Y, Nakazawa, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of hydration on the stability of the collagen-like triple-helical structure of [4(R)-hydroxyprolyl-4(R)-hydroxyprolylglycine]10
Biochemistry, 44, 2005
|
|
1VA2
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA3
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1WPK
 
 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | Descriptor: | ADA regulatory protein, ZINC ION | | Authors: | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2004-09-07 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
1WKI
 
 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
