8WM0
 
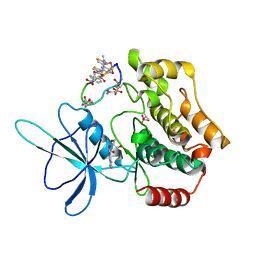 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
1NBK
 
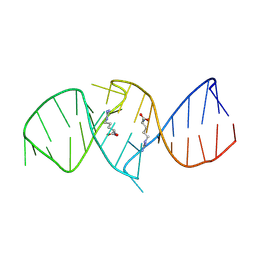 | | The structure of RNA aptamer for HIV Tat complexed with two argininamide molecules | | Descriptor: | 2-AMINO-5-GUANIDINO-PENTANOIC ACID, RNA aptamer | | Authors: | Matsugami, A, Kobayashi, S, Ouhashi, K, Uesugi, S, Yamamoto, R, Taira, K, Nishikawa, S, Kumar, P.K.R, Katahira, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2024-09-18 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Structure, 11, 2003
|
|
6J0O
 
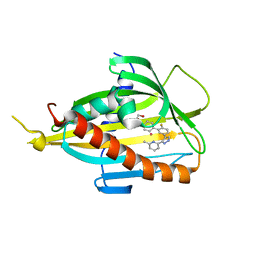 | | Crystal structure of CERT START domain in complex with compound SC1 | | Descriptor: | 2-[4-[2-fluoranyl-5-[3-(6-methylpyridin-2-yl)-1~{H}-pyrazol-4-yl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT, UNKNOWN ATOM OR ION | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-12-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6IEZ
 
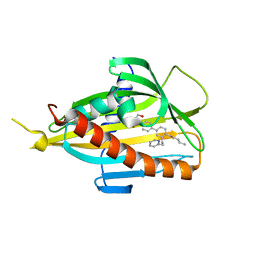 | | Crystal structure of CERT START domain in complex with compound B16 | | Descriptor: | 2-({4'-pentyl-3'-[2-(pyridin-2-yl)ethyl][1,1'-biphenyl]-4-yl}sulfonyl)ethan-1-ol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6J81
 
 | | Crystal structure of CERT START domain in complex with compound E14 | | Descriptor: | 2-[4-[4-propyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6IF0
 
 | | Crystal structure of CERT START domain in complex with compound D16 | | Descriptor: | 2-({4'-pentyl-3'-[(Z)-2-(pyridin-2-yl)ethenyl][1,1'-biphenyl]-4-yl}sulfonyl)ethan-1-ol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYL
 
 | | Crystal structure of CERT START domain in complex with compound E25A | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYI
 
 | | Crystal structure of CERT START domain in complex with compound E16 | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYJ
 
 | | Crystal structure of CERT START domain in complex with compound E16A | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYH
 
 | | Crystal structure of CERT START domain in complex with compound E5 | | Descriptor: | 2-[4-[3-~{tert}-butyl-5-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYK
 
 | | Crystal structure of CERT START domain in complex with compound E25 | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYG
 
 | | Crystal structure of CERT START domain in complex with compound B5 | | Descriptor: | 2-[4-[3-~{tert}-butyl-5-(2-pyridin-2-ylethyl)phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYM
 
 | | Crystal structure of CERT START domain in complex with compound E25B | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
3ABE
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a tetragonal crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
3ABD
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a monoclinic crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
7VMY
 
 | | Crystal structure of LimF prenyltransferase bound with GSPP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VMW
 
 | | Crystal structure of LimF prenyltransferase bound with a peptide substrate and GSPP | | Descriptor: | GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
2DB3
 
 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
