1Q66
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-6-AMINOMETHYL-8-phenylsulfanylmethyl-3H-QUINAZOLIN-4-ONE crystallized at pH 5.5 | | Descriptor: | 2-AMINO-6-AMINOMETHYL-8-PHENYLSULFANYLMETHYL-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
5AHG
 
 | | Thrombin in complex with ((4-chlorophenyl)sulfamoyl))diemethylamine | | Descriptor: | ((4-Chlorophenyl)sulfamoyl))dimethylamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-02-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
1Q65
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-DIAMINO-8-(2-dimethylaminoethylsulfanylmethyl)-3H-QUINAZOLIN-4-ONE crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(2-DIMETHYLAMINOETHYLSULFANYLMETHYL)-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
4Y3Q
 
 | |
4Y3D
 
 | |
4Y4Z
 
 | | Endothiapepsin in complex with fragment 73 | | Descriptor: | (1Z)-N,N-diethylethanimidamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y58
 
 | |
4Y5K
 
 | | Endothiapepsin in complex with fragment 274 | | Descriptor: | (1S)-2-amino-1-(4-fluorophenyl)ethanol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
1Q63
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-Diamino-8-(1H-imidazol-2-ylsulfanylmethyl)-3H-quinazoline-4-one crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
4YD5
 
 | | Endothiapepsin in complex with fragment 236 | | Descriptor: | 1-(2-chloro-5-nitrophenyl)-N-methylmethanamine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.209 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4YD7
 
 | | Endothiapepsin in complex with fragment 255 | | Descriptor: | 2-(imidazo[1,2-a]pyridin-2-yl)-N-phenylacetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y5L
 
 | | Endothiapepsin in its apo form | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4YCT
 
 | |
4YD3
 
 | | Endothiapepsin in complex with fragment 224 | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.248 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y50
 
 | |
5AFZ
 
 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-(2-((4- carbamimidoylphenyl)methylamino)-2-oxo-propyl)-3-phenyl-propanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, N-(BENZYLSULFONYL)-D-PHENYLALANYL-N-(4-CARBAMIMIDOYLBENZYL)GLYCINAMIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-01-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
1Q4W
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE | | Descriptor: | 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1R5Y
 
 | | Crystal Structure of TGT in complex with 2,6-Diamino-3H-Quinazolin-4-one Crystallized at PH 5.5 | | Descriptor: | 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Garcia, G.A, Stubbs, M.T, Klebe, G. | | Deposit date: | 2003-10-13 | | Release date: | 2004-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
8BJL
 
 | |
4E7R
 
 | | Thrombin in complex with 3-amidinophenylalanine inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-{[(2',4'-dichlorobiphenyl-3-yl)sulfonyl]amino}-3-oxopropyl]benzenecarboximidamide, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2012-03-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New 3-amidinophenylalanine-derived inhibitors of matriptase
MEDCHEMCOMM, 3, 2012
|
|
4Q4Q
 
 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 2-[(Thiophen-2-ylmethyl)amino]-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 2-[(thiophen-2-ylmethyl)amino]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Beyond Affinity: Enthalpy-Entropy Factorization Unravels Complexity of a Flat Structure-Activity Relationship for Inhibition of a tRNA-Modifying Enzyme.
J.Med.Chem., 57, 2014
|
|
3KIG
 
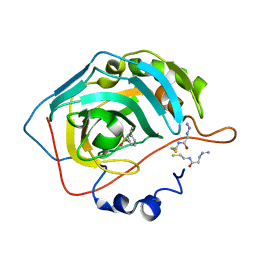 | | Mutant carbonic anhydrase II in complex with an azide and an alkyne | | Descriptor: | 2-azido-N-(2-sulfanylethyl)ethanamide, 3-ethynylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Schulze-Wischeler, J, Niehage, N.U, Heine, A, Klebe, G. | | Deposit date: | 2009-11-02 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Stereo- and Regioselective Azide/Alkyne Cycloadditions in Carbonic Anhydrase II via Tethering, Monitored by Crystallography and Mass Spectrometry.
Chemistry, 17, 2011
|
|
6I2B
 
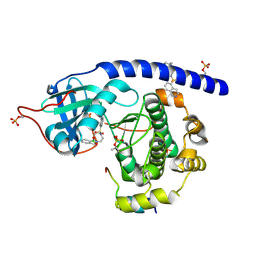 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Cricetulus Griseus in complex with compounds RKp153 and RKp117 | | Descriptor: | UPF0418 protein FAM164A, [2-[(4-isoquinolin-5-ylsulfonyl-1,4-diazepan-1-yl)methyl]phenyl]boronic acid, beta-D-ribopyranose, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-15 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
3LJO
 
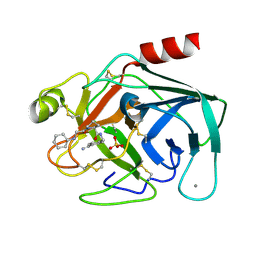 | | Bovine trypsin in complex with UB-THR 11 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Wegscheid-Gerlach, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties.
J.Mol.Biol., 405, 2011
|
|
3LQG
 
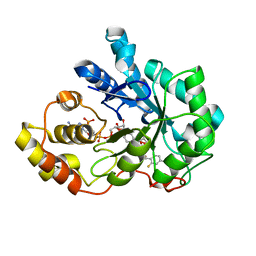 | | Human aldose reductase mutant T113A complexed with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-02-09 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
