1WE0
 
 | | Crystal structure of peroxiredoxin (AhpC) from Amphibacillus xylanus | | Descriptor: | AMMONIUM ION, alkyl hydroperoxide reductase C | | Authors: | Kitano, K, Kita, A, Hakoshima, T, Niimura, Y, Miki, K. | | Deposit date: | 2004-05-21 | | Release date: | 2005-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus
Proteins, 59, 2005
|
|
7E8R
 
 | | EcoT38I restriction endonuclease | | Descriptor: | EcoT38I restriction endonuclease, GLYCEROL | | Authors: | Kita, K, Mikami, B. | | Deposit date: | 2021-03-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of EcoT38I restriction endonuclease
To Be Published
|
|
7EDB
 
 | | EcoT38I restriction endonuclease complexed with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kita, K, Mikami, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural analysis of EcoT38I restriction endonuclease
To Be Published
|
|
1GEH
 
 | | CRYSTAL STRUCTURE OF ARCHAEAL RUBISCO (RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE) | | Descriptor: | RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, SULFATE ION | | Authors: | Kitano, K, Maeda, N, Fukui, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2000-11-13 | | Release date: | 2001-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Novel-Type Archaeal Rubisco with Pentagonal Symmetry
Structure, 9, 2001
|
|
5BRO
 
 | | Crystal structure of modified HexB (modB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase subunit beta, FORMIC ACID, ... | | Authors: | Kitakaze, K, Maita, N, Itoh, K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protease-resistant modified human beta-hexosaminidase B ameliorates symptoms in GM2 gangliosidosis model.
J.Clin.Invest., 126, 2016
|
|
2SFA
 
 | | SERINE PROTEINASE FROM STREPTOMYCES FRADIAE ATCC 14544 | | Descriptor: | SERINE PROTEINASE | | Authors: | Kitadokoro, K, Tsuzuki, H. | | Deposit date: | 1994-04-25 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Purification, characterization, primary structure, crystallization and preliminary crystallographic study of a serine proteinase from Streptomyces fradiae ATCC 14544.
Eur.J.Biochem., 220, 1994
|
|
1G8Q
 
 | | CRYSTAL STRUCTURE OF HUMAN CD81 EXTRACELLULAR DOMAIN, A RECEPTOR FOR HEPATITIS C VIRUS | | Descriptor: | CD81 ANTIGEN, EXTRACELLULAR DOMAIN | | Authors: | Kitadokoro, K, Bolognesi, M, Bordo, D, Grandi, G, Galli, G, Petracca, R, Falugi, F. | | Deposit date: | 2000-11-20 | | Release date: | 2001-02-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs.
EMBO J., 20, 2001
|
|
1IV5
 
 | | New Crystal Form of Human CD81 Large Extracellular Loop. | | Descriptor: | CD81 antigen | | Authors: | Kitadokoro, K, Bolognesi, M, Grandi, G, Marco, P, Galli, G, Petracca, R, Fabiana, F. | | Deposit date: | 2002-03-14 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Subunit Association and Conformational Flexibility in the Head-subdomain of Human CD81 Large Extracellular Loop.
Biol.Chem., 383, 2002
|
|
6KSL
 
 | | Staphylococcus aureus lipase - S116A inactive mutant | | Descriptor: | CALCIUM ION, LAURIC ACID, Lipase 2, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
6AID
 
 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | Descriptor: | CALCIUM ION, Esterase, LACTIC ACID, ... | | Authors: | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
6KSM
 
 | | Staphylococcus aureus lipase -Orlistat complex | | Descriptor: | (2S,3S,5S)-5-[(N-FORMYL-L-LEUCYL)OXY]-2-HEXYL-3-HYDROXYHEXADECANOIC ACID, CALCIUM ION, LAURIC ACID, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
3AM2
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
6KSI
 
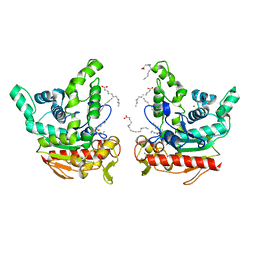 | | Staphylococcus aureus lipase - native | | Descriptor: | CALCIUM ION, HEXANOIC ACID, LAURIC ACID, ... | | Authors: | Kitadokoro, K, Tanaka, M, Kamitani, S. | | Deposit date: | 2019-08-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat.
Sci Rep, 10, 2020
|
|
2EBF
 
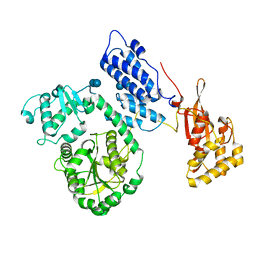 | |
2E1E
 
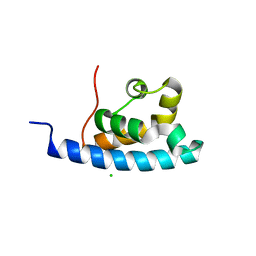 | | Crystal structure of the HRDC Domain of Human Werner Syndrome Protein, WRN | | Descriptor: | CHLORIDE ION, Werner syndrome ATP-dependent helicase | | Authors: | Kitano, K, Yoshihara, N, Hakoshima, T. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the HRDC domain of human Werner syndrome protein, WRN
J.Biol.Chem., 282, 2007
|
|
2EBH
 
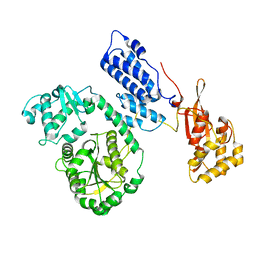 | |
2E1F
 
 | | Crystal structure of the HRDC Domain of Human Werner Syndrome Protein, WRN | | Descriptor: | CHLORIDE ION, Werner syndrome ATP-dependent helicase | | Authors: | Kitano, K, Yoshihara, N, Hakoshima, T. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HRDC domain of human Werner syndrome protein, WRN
J.Biol.Chem., 282, 2007
|
|
2D2Q
 
 | |
2EC5
 
 | |
3AAF
 
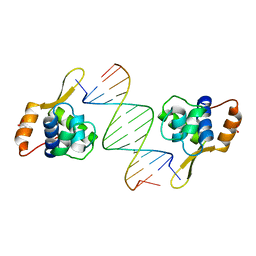 | | Structure of WRN RQC domain bound to double-stranded DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*CP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*GP*T)-3'), Werner syndrome ATP-dependent helicase | | Authors: | Kitano, K, Hakoshima, T. | | Deposit date: | 2009-11-16 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN
Structure, 18, 2010
|
|
3VIS
 
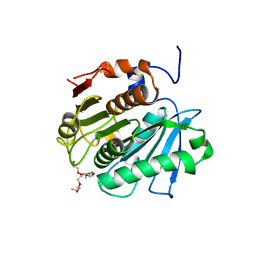 | | Crystal structure of cutinase Est119 from Thermobifida alba AHK119 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Esterase | | Authors: | Kitadokoro, K, Thumarat, U, Nakamura, R, Nishimura, K, Karatani, H, Suzuki, H, Kawai, F. | | Deposit date: | 2011-10-11 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of cutinase Est119 from Thermobida alba AHK119 that can degrade modpolyethylene terephthalate at 1.76 A resolution.
POLYM.DEGRAD.STAB., 97, 2012
|
|
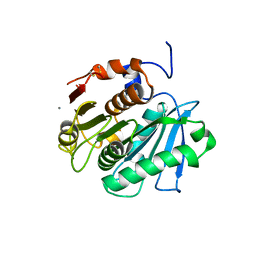
3WYN
 
 | |
5GN6
 
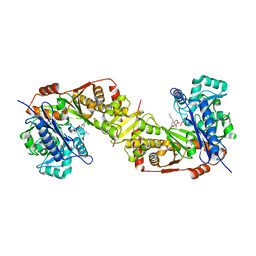 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17b | | Descriptor: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5GN7
 
 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
4JDB
 
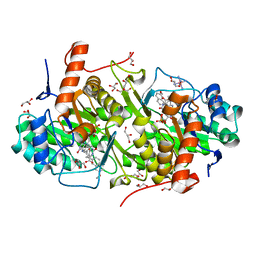 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-005 | | Descriptor: | 5-[2-(7-methoxynaphthalen-2-yl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-005
To be Published
|
|
