9IWQ
 
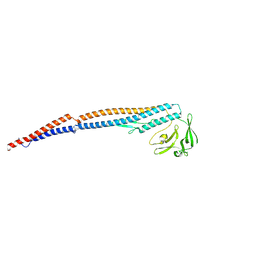 | | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament | | Descriptor: | Flagellin | | Authors: | Waraich, K, Makino, F, Miyata, T, Kinoshita, M, Minamino, T, Namba, K. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament
To Be Published
|
|
2RU7
 
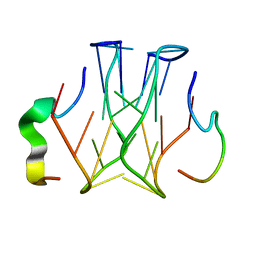 | | Refined structure of RNA aptamer in complex with the partial binding peptide of prion protein | | Descriptor: | P16 peptide from Major prion protein, RNA_(5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3') | | Authors: | Hayashi, T, Oshima, H, Mashima, T, Nagata, T, Katahira, M, Kinoshita, M. | | Deposit date: | 2013-12-24 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding of an RNA aptamer and a partial peptide of a prion protein: crucial importance of water entropy in molecular recognition.
Nucleic Acids Res., 42, 2014
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7CTN
 
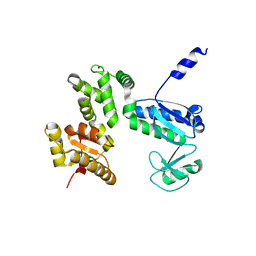 | | Structure of the 328-692 fragment of FlhA (E351A/D356A) | | Descriptor: | Flagellar biosynthesis protein FlhA | | Authors: | Kida, M, Takekawa, N, Kinoshita, M, Inoue, Y, Minamino, T, Imada, K. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The FlhA linker mediates flagellar protein export switching during flagellar assembly.
Commun Biol, 4, 2021
|
|
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
5B0O
 
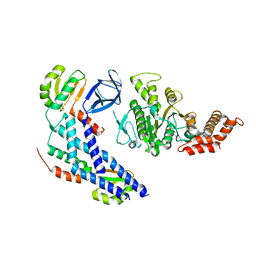 | | Structure of the FliH-FliI complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Flagellar assembly protein FliH, Flagellum-specific ATP synthase | | Authors: | Imada, K, Uchida, Y, Kinoshita, M, Namba, K, Minamino, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6KFQ
 
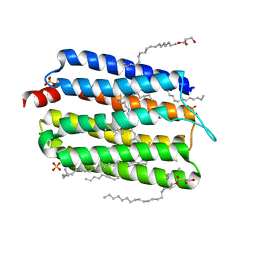 | | Crystal structure of thermophilic rhodopsin from Rubrobacter xylanophilus | | Descriptor: | RETINAL, Rhodopsin, SULFATE ION, ... | | Authors: | Suzuki, K, Akiyama, T, Hayashi, T, Yasuda, S, Kanehara, K, Kojima, K, Tanabe, M, Kato, R, Senda, T, Sudo, Y, Kinoshita, M, Murata, T. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Does a Microbial Rhodopsin RxR Realize Its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained?
J.Phys.Chem.B, 124, 2020
|
|
6K84
 
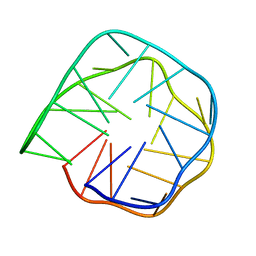 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
3AJC
 
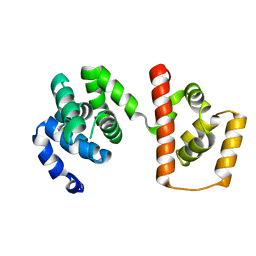 | | Structure of the MC domain of FliG (PEV), a CW-biased mutant | | Descriptor: | Flagellar motor switch protein fliG | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the rotational switching mechanism of the bacterial flagellar motor
Plos Biol., 9, 2011
|
|
5YC8
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) | | Descriptor: | MERCURY (II) ION, Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKB
 
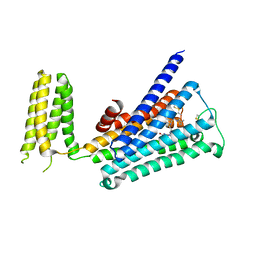 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
3A7M
 
 | | Structure of FliT, the flagellar type III chaperone for FliD | | Descriptor: | Flagellar protein fliT | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ADY
 
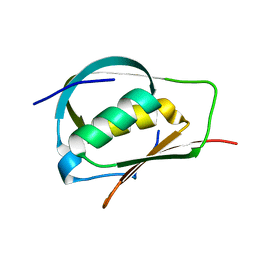 | | Crystal structure of DotD from Legionella | | Descriptor: | DotD | | Authors: | Imada, K, Nakano, N, Kubori, T, Kinoshita, M, Nagai, H. | | Deposit date: | 2010-01-29 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems
Plos Pathog., 6, 2010
|
|
6AI2
 
 | |
3VYB
 
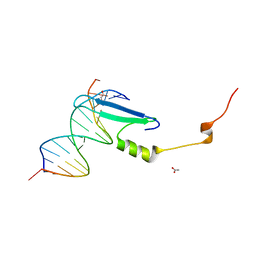 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/hmCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5HC)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3W1C
 
 | |
3W1D
 
 | |
3VXX
 
 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/5mCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5CM)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VYQ
 
 | | Crystal structure of the methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence in space group P1 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*AP*TP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
6AI1
 
 | |
6AI3
 
 | |
3VXV
 
 | | Crystal structure of methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*CP*(5CM)P*GP*GP*AP*CP*A)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
