4MNU
 
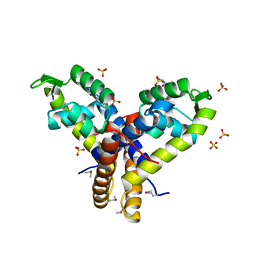 | | Crystal Structure of Uncharacterized SlyA-like Transcription Regulator from Listeria monocytogenes | | Descriptor: | GLYCEROL, SULFATE ION, SlyA-like transcription regulator | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Uncharacterized SlyA-like Transcription Regulator from
Listeria monocytogenes
To be Published
|
|
4MY3
 
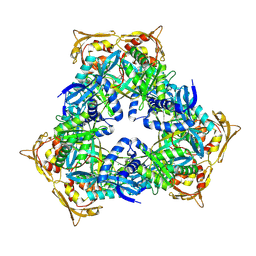 | | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GCN5-related N-acetyltransferase | | Authors: | Kim, Y, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida
To be Published
|
|
1T57
 
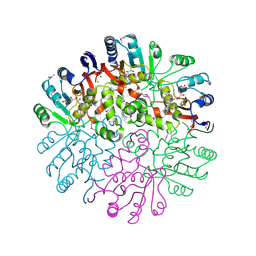 | | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum | | Descriptor: | Conserved Protein MTH1675, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION | | Authors: | Kim, Y, Joachimiak, A, Saridakis, V, Xu, X, Arrowsmith, C.H, Christendat, D, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum
To be Published
|
|
1SS4
 
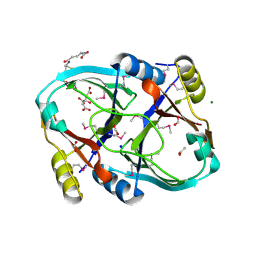 | |
1T07
 
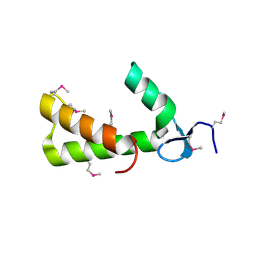 | | Crystal Structure of Conserved Protein of Unknown Function PA5148 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical UPF0269 protein PA5148 | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of PA5148 from Pseudomonas aeruginosa
To be Published
|
|
1TAQ
 
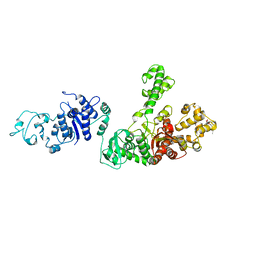 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
7V6U
 
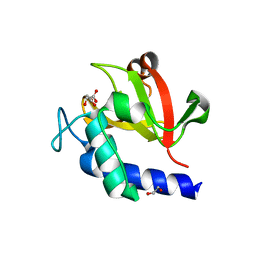 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | The crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6T
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6S
 
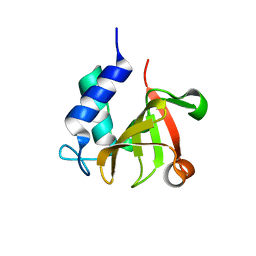 | | Crystal structure of bacterial peptidase | | Descriptor: | Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | The crystal structure fre
To be published
|
|
7EWR
 
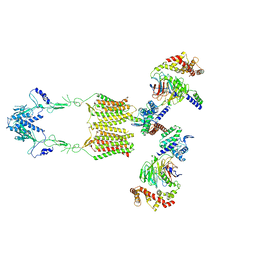 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:2:2 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7EWP
 
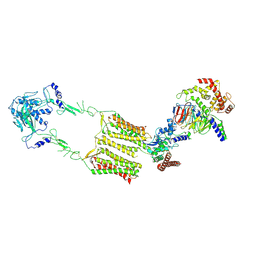 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:1:1 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7CAZ
 
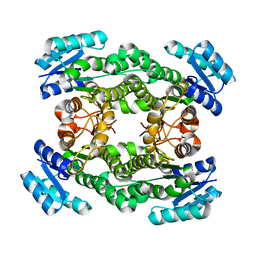 | | Crystal structure of bacterial reductase | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, GLYCEROL | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7CAX
 
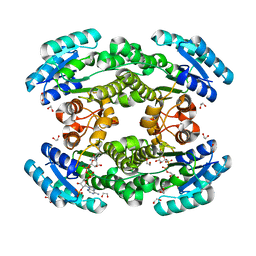 | | Crystal structure of bacterial reductase | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-ACP reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7CAW
 
 | | Crystal structure of bacterial reductase | | Descriptor: | 3-oxoacyl-ACP reductase FabG, GLYCEROL | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7F27
 
 | | Crystal structure of polyketide ketosynthase | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
1YB4
 
 | | Crystal Structure of the Tartronic Semialdehyde Reductase from Salmonella typhimurium LT2 | | Descriptor: | tartronic semialdehyde reductase | | Authors: | Kim, Y, Wu, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
9J2F
 
 | | Structure of photosynthetic LH1-RC complex from the purple bacterium Blastochloris tepida | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 15-cis-1,2-dihydroneurosporene, Antenna complex alpha/beta subunit domain-containing protein, ... | | Authors: | Kimura, Y, Kanno, R, Mori, K, Matsuda, Y, Seto, R, Takenaka, S, Mino, H, Ohkubo, T, Honda, M, Sasaki, Y.C, Kishikawa, J, Mitsuoka, K, Mio, K, Hall, M, Purba, E.R, Mochizuki, T, Mizoguchi, A, Humbel, B.M, Madigan, M.T, Wang-Otomo, Z.-Y, Tani, K. | | Deposit date: | 2024-08-06 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The thermal-stable LH1-RC complex of a hot spring purple bacterium powers photosynthesis with extremely low-energy near-infrared light.
To Be Published
|
|
1AT9
 
 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
1TX6
 
 | | trypsin:BBI complex | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Song, H.K, Park, E.Y, Kim, J.A, Kim, H.W, Kim, Y.S. | | Deposit date: | 2004-07-02 | | Release date: | 2005-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin
J.Mol.Biol., 343, 2004
|
|
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
1T52
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
2OJN
 
 | |
2OJO
 
 | |
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
