5HPF
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain from Acinetobacter | | 分子名称: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-09-07 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.309 Å) | | 主引用文献 | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
7TRV
 
 | | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Kim, Y, Tesar, C, Crawford, M, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-31 | | 公開日 | 2022-02-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
6W2Z
 
 | | Crystal Structure of the Beta Lactamase Class A PenP from Bacillus subtilis in the Complex with the Non-beta- lactam Beta-lactamase Inhibitor Avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-08 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of the Beta Lactamase Class A PenP from Bacillus subtilis in the Complex with the Non-beta- lactam Beta-lactamase Inhibitor Avibactam
To Be Published
|
|
7TMU
 
 | | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-20 | | 公開日 | 2022-02-02 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis
To Be Published
|
|
7TRW
 
 | | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | 分子名称: | 3-HYDROXYBENZOIC ACID, LysR-family transcriptional regulatory protein, PHOSPHATE ION | | 著者 | Kim, Y, Tesar, C, Crawford, M, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-31 | | 公開日 | 2022-02-09 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7TKV
 
 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | 分子名称: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-17 | | 公開日 | 2022-01-26 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
7THW
 
 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | 分子名称: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | 著者 | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-12 | | 公開日 | 2022-01-26 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
7TJ1
 
 | | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Tesar, C, Pastore, T, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-14 | | 公開日 | 2022-01-26 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
To Be Published
|
|
6W01
 
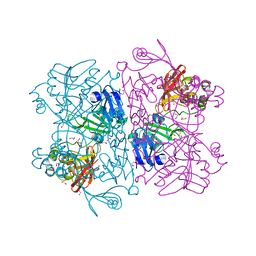 | | The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-28 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-29 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | 著者 | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-04 | | 公開日 | 2020-03-25 | | 最終更新日 | 2022-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
7TWC
 
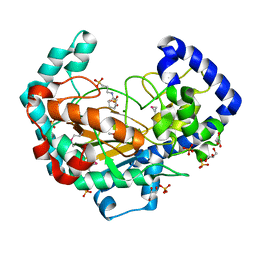 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DUF1479 domain-containing protein, GLYCEROL, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS
To Be Published
|
|
7TWE
 
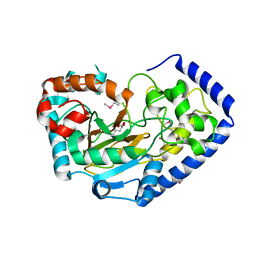 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DUF1479 domain-containing protein, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid
To Be Published
|
|
6W34
 
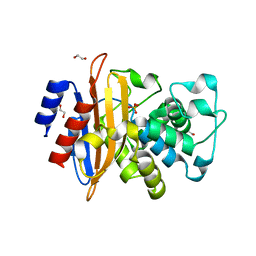 | | Crystal Structure of Class A Beta-lactamase from Bacillus cereus | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-08 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal Structure of Class A Beta-lactamase from
Bacillus cereus
To Be Published
|
|
6VXS
 
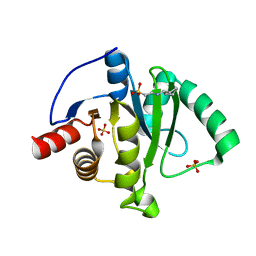 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Non-structural protein 3, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-24 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W33
 
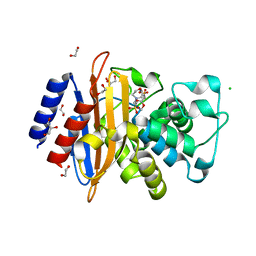 | | Crystal Structure of Class A Beta-lactamase from Bacillus cereus in the Complex with the Beta-lactamase Inhibitor Clavulanate | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-08 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Class A Beta-lactamase from
Bacillus cereus in the Complex with the Beta-lactamase Inhibitor Clavulanate
To Be Published
|
|
6W61
 
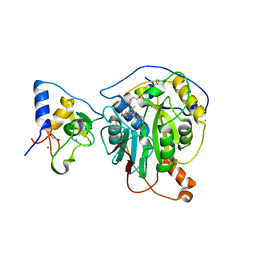 | | Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2. | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, CHLORIDE ION, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine
Biorxiv, 2020
|
|
3K0L
 
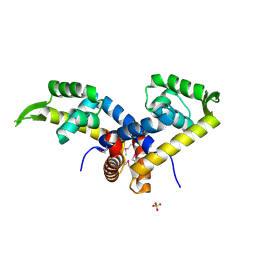 | | Crystal Structure of Putative MarR Family Transcriptional Regulator from Acinetobacter sp. ADP | | 分子名称: | Repressor protein, SULFATE ION | | 著者 | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-09-24 | | 公開日 | 2009-10-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.352 Å) | | 主引用文献 | Crystal Structure of Putative MarR Family Transcriptional Regulator from Acinetobacter sp. ADP
To be Published
|
|
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | 分子名称: | Fab S24-1379, heavy chain, light chain, ... | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-11-15 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7SUE
 
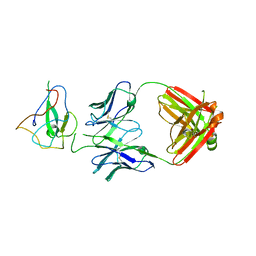 | | Crystal Structure of Human Fab S24-188 in the complex with the N-teminal Domain of Nucleocapsid protein from SARS CoV-2 | | 分子名称: | Nucleoprotein, S24-188 Fab Heavy chain, S24-188 Fab Light chain | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-11-17 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7SF6
 
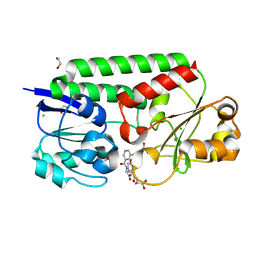 | | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense | | 分子名称: | 1,2-ETHANEDIOL, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Patel, H.P, Nordquist, K.A, Schaab, K.M, Sha, J, Babnigg, G, Bond, A.H, Joachimiak, A, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2021-10-03 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense
To Be Published
|
|
3KTB
 
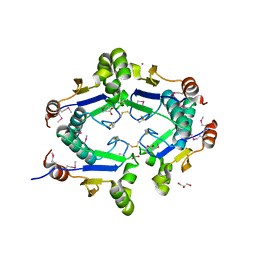 | | Crystal Structure of Arsenical Resistance Operon Trans-acting Repressor from Bacteroides vulgatus ATCC 8482 | | 分子名称: | ACETIC ACID, Arsenical resistance operon trans-acting repressor, CALCIUM ION, ... | | 著者 | Kim, Y, Tesar, C, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-11-24 | | 公開日 | 2009-12-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Arsenical Resistance Operon Trans-acting Repressor from Bacteroides vulgatus ATCC 8482
To be Published
|
|
3KV1
 
 | |
3KN3
 
 | | Crystal Structure of LysR Substrate Binding Domain (25-263) of Putative Periplasmic Protein from Wolinella succinogenes | | 分子名称: | ACETIC ACID, CITRIC ACID, GLUTATHIONE, ... | | 著者 | Kim, Y, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-11-11 | | 公開日 | 2009-12-01 | | 最終更新日 | 2011-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.412 Å) | | 主引用文献 | Crystal Structure of LysR Substrate Binding Domain from Wolinella succinogenes
To be Published
|
|
7CUM
 
 | | Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626 | | 分子名称: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | 著者 | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | 登録日 | 2020-08-23 | | 公開日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
