2PDO
 
 | | Crystal Structure of the Putative Acetyltransferase of GNAT Family from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acetyltransferase ypeA, ... | | Authors: | Kim, Y, Li, H, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-01 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Acetyltransferase of GNAT Family from Shigella flexneri
To be Published
|
|
2P13
 
 | |
2QM1
 
 | | Crystal structure of glucokinase from Enterococcus faecalis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glucokinase, ... | | Authors: | Kim, Y, Joachimiak, G, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of Glucokinase from Enterococcus faecalis.
To be Published
|
|
2B20
 
 | | Crystal Structure of Enterochelin Esterase from Shigella flexneri Enterochelin Esterase | | Descriptor: | L(+)-TARTARIC ACID, enterochelin esterase | | Authors: | Kim, Y, Maltseva, N, Dementieva, I, Quartey, P, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Enterochelin Esterase from Shigella flexneri
Enterochelin Esterase
To be Published
|
|
3CDK
 
 | | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis | | Descriptor: | Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit A, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B | | Authors: | Kim, Y, Zhou, M, Stols, L, Eschenfeldt, W, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis.
To be Published
|
|
2B81
 
 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Luciferase-like monooxygenase, ... | | Authors: | Kim, Y, Li, H, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus
To be Published
|
|
3CDH
 
 | | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi DSS-3 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Kim, Y, Volkart, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi.
To be Published
|
|
2QQZ
 
 | | Crystal structure of putative glyoxalase family protein from Bacillus anthracis | | Descriptor: | GLYCEROL, Glyoxalase family protein, putative, ... | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Putative Glyoxalase Family Protein from Bacillus anthracis.
To be Published
|
|
2B67
 
 | | Crystal structure of the Nitroreductase Family Protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | ACETIC ACID, COG0778: Nitroreductase, FLAVIN MONONUCLEOTIDE | | Authors: | Kim, Y, Volkart, L, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Nitroreductase Family Protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
2QQY
 
 | | Crystal structure of ferritin like, diiron-carboxylate proteins from Bacillus anthracis str. Ames | | Descriptor: | Sigma B operon | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Ferritin like, Diiron-carboxylate Proteins from Bacillus anthracis str. Ames.
To be Published
|
|
2RA5
 
 | | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor | | Descriptor: | ISOPROPYL ALCOHOL, Putative transcriptional regulator, S,R MESO-TARTARIC ACID | | Authors: | Kim, Y, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
2R6H
 
 | | Crystal structure of the domain comprising the NAD binding and the FAD binding regions of the NADH:ubiquinone oxidoreductase, Na translocating, F subunit from Porphyromonas gingivalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH:ubiquinone oxidoreductase, Na translocating, ... | | Authors: | Kim, Y, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Domain Comprising the Regions Binding NAD and FAD from the NADH:Ubiquinone Oxidoreductase, Na Translocating, F Subunit from Porphyromonas gingivalis.
To be Published
|
|
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | Fab S24-1379, heavy chain, light chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7SUE
 
 | | Crystal Structure of Human Fab S24-188 in the complex with the N-teminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | Nucleoprotein, S24-188 Fab Heavy chain, S24-188 Fab Light chain | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
3QOD
 
 | | Crystal Structure of Heterocyst Differentiation Protein, HetR from Fischerella mv11 | | Descriptor: | Heterocyst differentiation protein | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3R3T
 
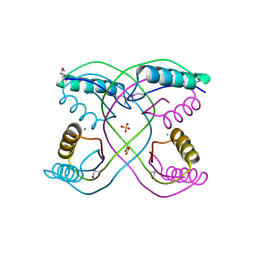 | | Crystal Structure of 30S Ribosomal Protein S from Bacillus anthracis | | Descriptor: | 30S ribosomal protein S6, CALCIUM ION, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of 30S Ribosomal Protein S from Bacillus anthracis
To be Published
|
|
3RE3
 
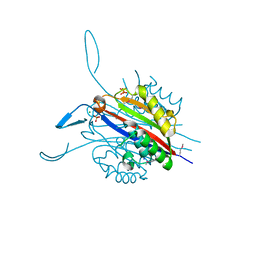 | | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase from Francisella tularensis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase from Francisella tularensis
To be Published
|
|
3RJU
 
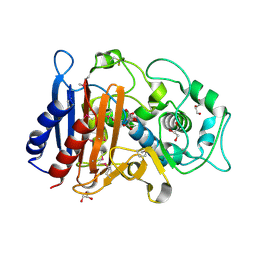 | | Crystal Structure of Beta-lactamase/D-alanine Carboxypeptidase from Yersinia pestis complexed with citrate | | Descriptor: | Beta-lactamase/D-alanine Carboxypeptidase, CITRIC ACID, GLYCEROL | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Beta-lactamase/D-alanine Carboxypeptidase from Yersinia pestis complexed with citrate
To be Published
|
|
7TCB
 
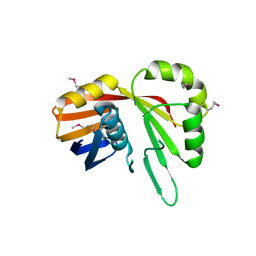 | | Crystal Structure of the YaeQ Family Protein VPA0551 from Vibrio parahaemolyticus | | Descriptor: | YaeQ family protein VPA0551 | | Authors: | Kim, Y, Mulligan, R, Maltseva, N, Gu, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the YaeQ Family Protein VPA0551 from Vibrio parahaemolyticus
To Be Published
|
|
7TEM
 
 | | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis
To Be Published
|
|
7TE5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis | | Descriptor: | MAGNESIUM ION, Pirin family protein Yhak | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis
To Be Published
|
|
7TG5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the Presence of Fe Ion from Yersinia pestis | | Descriptor: | CHLORIDE ION, FE (III) ION, Pirin family protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the presence of Fe ion from Yersinia pestis
To Be Published
|
|
7TFQ
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, FORMIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis
To Be Published
|
|
7THW
 
 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | Authors: | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-12 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
