5VUG
 
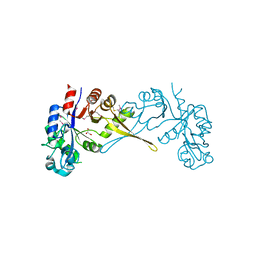 | | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase Domain of Uncharacterized Protein Rv2277c from Mycobacterium tuberculosis | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein Rv2277c | | Authors: | Kim, Y, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-14 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase Domain of Uncharacterized Protein Rv2277c from Mycobacterium tuberculosis
To Be Published
|
|
5VVI
 
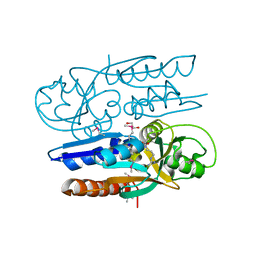 | | Crystal Structure of the Ligand Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens in the Complex with Octopine | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
9BZB
 
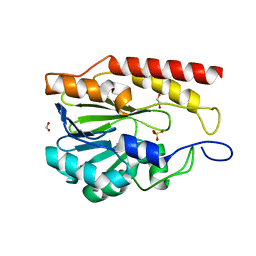 | | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2
To Be Published
|
|
2G7G
 
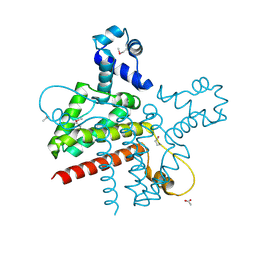 | | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1 | | Descriptor: | ACETIC ACID, Rha04620, Putative Transcriptional Regulator | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1
To be Published
|
|
6DFP
 
 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6DEB
 
 | | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional protein FolD, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni
To Be Published
|
|
6DE8
 
 | | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase from Campylobacter jejuni | | Descriptor: | Bifunctional protein FolD, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Zhang, R, Peterson, S.N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-30 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase from Campylobacter jejuni
To Be Published
|
|
3HBC
 
 | | Crystal Structure of Choloylglycine Hydrolase from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, Choloylglycine hydrolase, GLYCEROL | | Authors: | Kim, Y, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Crystal Structure of Choloylglycine Hydrolase from Bacteroides thetaiotaomicron VPI
To be Published
|
|
3HJJ
 
 | | Crystal Structure of Maltose O-acetyltransferase from Bacillus anthracis | | Descriptor: | ACETIC ACID, GLYCEROL, Maltose O-acetyltransferase, ... | | Authors: | Kim, Y, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Crystal Structure of Maltose O-acetyltransferase from Bacillus anthracis
To be Published
|
|
1MKI
 
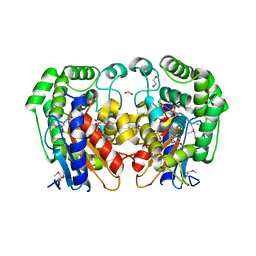 | | Crystal Structure of Bacillus Subtilis Probable Glutaminase, APC1040 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable Glutaminase ybgJ | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-06-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2FSW
 
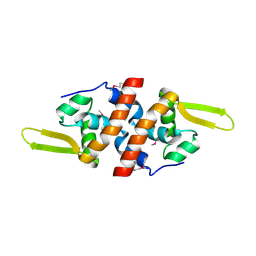 | | Crystal Structure of the Putative Transcriptional Regualator, MarR family from Porphyromonas gingivalis W83 | | Descriptor: | PG_0823 protein | | Authors: | Kim, Y, Quartey, P, Buelt, J, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Transcriptional Regualator, MarR family from Porphyromonas gingivalis W83
To be Published
|
|
2FSR
 
 | | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, acetyltransferase | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Gu, J, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58
To be Published
|
|
5L07
 
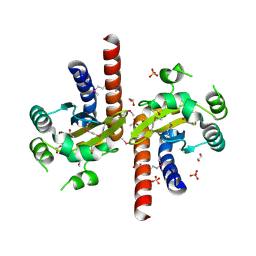 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Quorum-sensing transcriptional activator, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5KWS
 
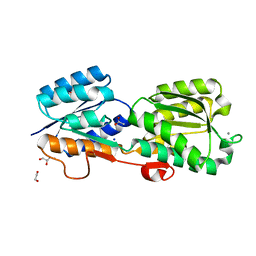 | | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose
To Be Published
|
|
5L09
 
 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica in complex with 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, ACETIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5KXJ
 
 | | Crystal Structure of L-Aspartate Oxidase from Salmonella typhimurium in the Complex with Substrate L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Osipiuk, J, Mulligan, R, Makowska-Grzyska, M, Maltseva, N, Shatsman, S, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of L-Aspartate Oxidase from Salmonella typhimurium in the Complex with Substrate L-Aspartate
To Be Published
|
|
5L10
 
 | | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winan, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia.
To Be Published
|
|
7RSK
 
 | | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein | | Authors: | Kim, Y, Nocek, B, Endres, M, Joachimiak, G, Johnson, J, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus
To Be Published
|
|
8EP7
 
 | | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in complex with NADP | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase (NADP(+)) 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Maltseva, N, Osipiuk, J, Gu, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in the complex with NADP.
To Be Published
|
|
2VRC
 
 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | Descriptor: | TRIPHENYLMETHANE REDUCTASE | | Authors: | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
7TOC
 
 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | Authors: | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
2VRB
 
 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIPHENYLMETHANE REDUCTASE | | Authors: | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
4Q63
 
 | | Crystal Structure of Legionella Uncharacterized Protein Lpg0364 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Crystal Structure of Legionella Uncharacterized Protein Lpg0364
To be Published
|
|
1XPP
 
 | | Crystal Structure of TA1416,DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum | | Descriptor: | ACETIC ACID, DNA-directed RNA polymerase subunit L, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of TA1416, DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum
To be Published
|
|
3EFB
 
 | | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri | | Descriptor: | ACETIC ACID, Probable sor-operon regulator | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri
To be Published
|
|
