4N5L
 
 | |
4N5N
 
 | |
3VLE
 
 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLD
 
 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2012-04-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLF
 
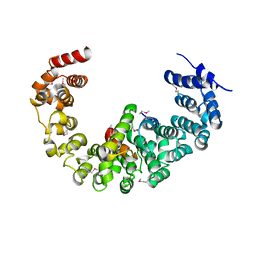 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
4B5Q
 
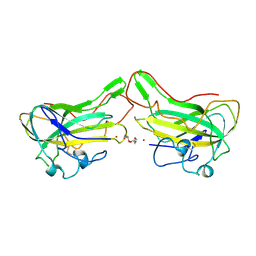 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | 分子名称: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | 著者 | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | 登録日 | 2012-08-07 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
4O9C
 
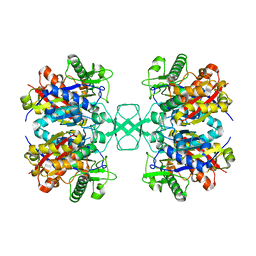 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | 分子名称: | Acetyl-CoA acetyltransferase, COENZYME A | | 著者 | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | 登録日 | 2014-01-02 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4O9A
 
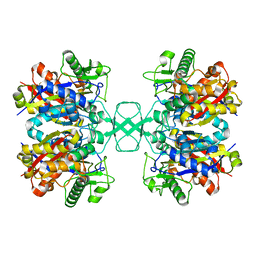 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | 分子名称: | Acetyl-CoA acetyltransferase | | 著者 | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | 登録日 | 2014-01-02 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4O99
 
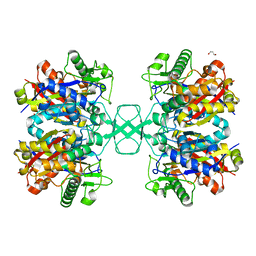 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | 分子名称: | Acetyl-CoA acetyltransferase, GLYCEROL | | 著者 | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | 登録日 | 2014-01-02 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
1MP6
 
 | |
3GWJ
 
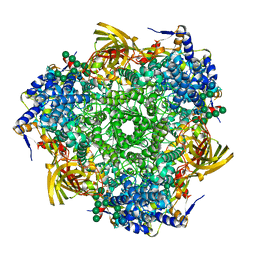 | | Crystal structure of Antheraea pernyi arylphorin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | 著者 | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | 登録日 | 2009-04-01 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
1P68
 
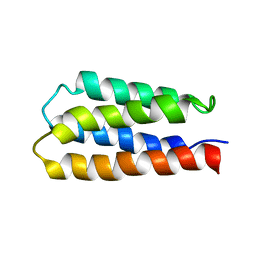 | | Solution structure of S-824, a de novo designed four helix bundle | | 分子名称: | De novo designed protein S-824 | | 著者 | Wei, Y, Kim, S, Fela, D, Baum, J, Hecht, M.H. | | 登録日 | 2003-04-29 | | 公開日 | 2003-11-11 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a de novo protein from a designed combinatorial library.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
3HQD
 
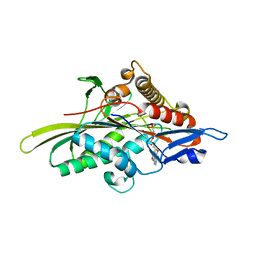 | | Human kinesin Eg5 motor domain in complex with AMPPNP and Mg2+ | | 分子名称: | Kinesin-like protein KIF11, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Parke, C.L, Wojcik, E.J, Kim, S, Worthylake, D.K. | | 登録日 | 2009-06-05 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | ATP Hydrolysis in Eg5 Kinesin Involves a Catalytic Two-water Mechanism.
J.Biol.Chem., 285, 2010
|
|
3OU0
 
 | | re-refined 3CS0 | | 分子名称: | Periplasmic serine endoprotease DegP, heptapeptide, pentapeptide | | 著者 | Sauer, R.T, Grant, R.A, Kim, S. | | 登録日 | 2010-09-14 | | 公開日 | 2011-01-19 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Cell(Cambridge,Mass.), 145, 2011
|
|
6MW7
 
 | | Crystal structure of ATPase module of SMCHD1 bound to ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Pedersen, L.C, Inoue, K, Kim, S, Perera, L, Shaw, N.D. | | 登録日 | 2018-10-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | A ubiquitin-like domain is required for stabilizing the N-terminal ATPase module of human SMCHD1.
Commun Biol, 2, 2019
|
|
2O0O
 
 | | Crystal structure of TL1A | | 分子名称: | MAGNESIUM ION, TNF superfamily ligand TL1A | | 著者 | Jin, T.C, Kim, S, Guo, F, Howard, A.J, Zhang, Y.Z. | | 登録日 | 2006-11-27 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-ray crystal structure of TNF ligand family member TL1A at 2.1A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
1PUL
 
 | | Solution structure for the 21KDa caenorhabditis elegans protein CE32E8.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR33 | | 分子名称: | Hypothetical protein C32E8.3 in chromosome I | | 著者 | Tejero, R, Aramini, J.M, Swapna, G.V.T, Monleon, D, Chiang, Y, Macapagal, D, Gunsalus, K.C, Kim, S, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-06-25 | | 公開日 | 2005-06-21 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Backbone 1H, 15N and 13C assignments for the 21 kDa Caenorhabditis elegans homologue of "brain-specific" protein.
J.Biomol.Nmr, 28, 2004
|
|
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | 分子名称: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | 著者 | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | 登録日 | 2003-11-21 | | 公開日 | 2004-04-27 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
1TVI
 
 | | Solution structure of TM1509 from Thermotoga maritima: VT1, a NESGC target protein | | 分子名称: | Hypothetical UPF0054 protein TM1509 | | 著者 | Penhoat, C.H, Atreya, H.S, Kim, S, Li, Z, Yee, A, Xiao, R, Murray, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-06-29 | | 公開日 | 2005-01-04 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
4REP
 
 | | Crystal Structure of gamma-carotenoid desaturase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Gamma-carotene desaturase | | 著者 | Ahn, J.-W, Kim, E.-J, Kim, S, Kim, K.-J. | | 登録日 | 2014-09-23 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of 1'-OH-carotenoid 3,4-desaturase from Nonlabens dokdonensis DSW-6.
Enzyme.Microb.Technol., 77, 2015
|
|
7XGF
 
 | |
7XGE
 
 | |
7XGG
 
 | |
6MZP
 
 | |
6MZN
 
 | |
