5XHW
 
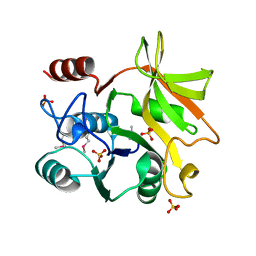 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
5XF2
 
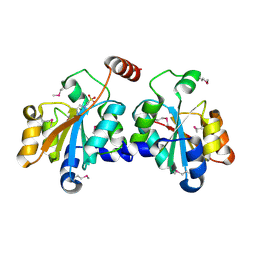 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7CPZ
 
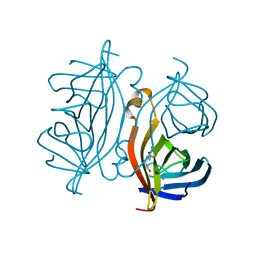 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | BIOTIN, Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CQ0
 
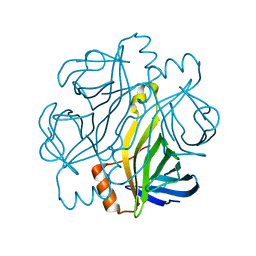 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
4B5Q
 
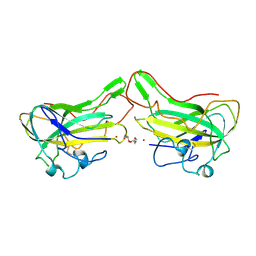 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | Descriptor: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | Authors: | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
4FZ5
 
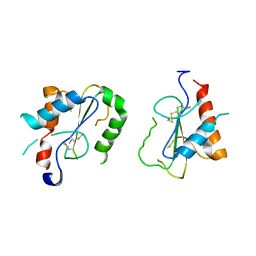 | | Crystal Structure of Human TIRAP TIR-domain | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Woo, J.R, Kim, S, Shoelson, S.E, Park, S. | | Deposit date: | 2012-07-06 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray Crystallographic Structure of TIR-Domain from the Human TIR-Domain Containing Adaptor Protein/MyD88 Adaptor-Like Protein (TIRAP/MAL)
Bull.Korean Chem.Soc., 33, 2013
|
|
2VTC
 
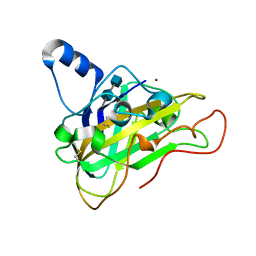 | | The structure of a glycoside hydrolase family 61 member, Cel61B from the Hypocrea jecorina. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CEL61B, NICKEL (II) ION | | Authors: | Karkehabadi, S, Hansson, H, Kim, S, Piens, K, Mitchinson, C, Sandgren, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The First Structure of a Glycoside Hydrolase Family 61 Member, Cel61B from the Hypocrea Jecorina, at 1.6 A Resolution.
J.Mol.Biol., 383, 2008
|
|
7CXY
 
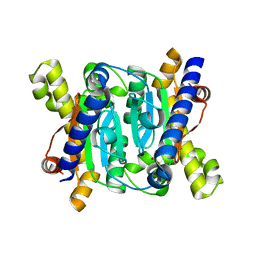 | | Structural insights into novel mechanisms of inhibition of the major b-carbonic anhydrase CafB from the pathogenic fungus Aspergillus fumigatus (zinc-bound form) | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Yeon, J, Sung, J, Kim, N.J, Hong, S. | | Deposit date: | 2020-09-02 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into novel mechanisms of inhibition of the major beta-carbonic anhydrase CafB from the pathogenic fungus Aspergillus fumigatus.
J.Struct.Biol., 213, 2021
|
|
7CXX
 
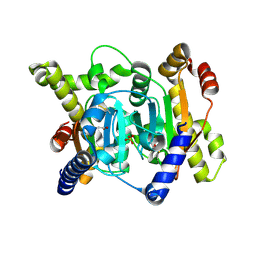 | | Structural insights into novel mechanisms of inhibition of the major b-carbonic anhydrase CafB from the pathogenic fungus Aspergillus fumigatus (disulfide-bonded form) | | Descriptor: | ACETATE ION, Carbonic anhydrase, SULFATE ION | | Authors: | Jin, M.S, Kim, S, Yeon, J, Sung, J, Kim, N.J, Hong, S. | | Deposit date: | 2020-09-02 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into novel mechanisms of inhibition of the major beta-carbonic anhydrase CafB from the pathogenic fungus Aspergillus fumigatus.
J.Struct.Biol., 213, 2021
|
|
7CXW
 
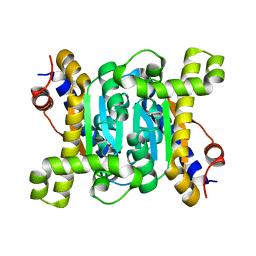 | | Structural insights into novel mechanisms of inhibition of the major b-carbonic anhydrase CafB from the pathogenic fungus Aspergillus fumigatus (C116 flipped form) | | Descriptor: | ACETATE ION, Carbonic anhydrase | | Authors: | Jin, M.S, Kim, S, Yeon, J, Sung, J, Kim, N.J, Hong, S. | | Deposit date: | 2020-09-02 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into novel mechanisms of inhibition of the major beta-carbonic anhydrase CafB from the pathogenic fungus Aspergillus fumigatus.
J.Struct.Biol., 213, 2021
|
|
5ZNX
 
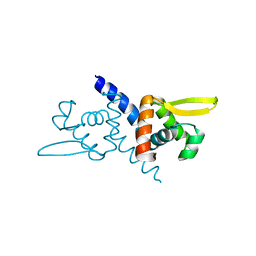 | | Crystal structure of CM14-treated HlyU from Vibrio vulnificus | | Descriptor: | Transcriptional activator | | Authors: | Park, N, Kim, S, Jo, I, Ahn, J, Hong, S, Jeong, S, Baek, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Small-molecule inhibitor of HlyU attenuates virulence of Vibrio species.
Sci Rep, 9, 2019
|
|
1I6A
 
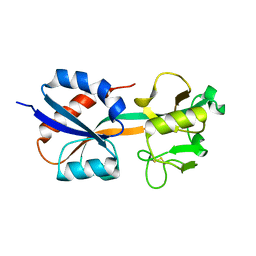 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF OXYR | | Descriptor: | HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I69
 
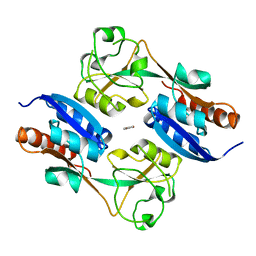 | | CRYSTAL STRUCTURE OF THE REDUCED FORM OF OXYR | | Descriptor: | BENZOIC ACID, HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
3ATT
 
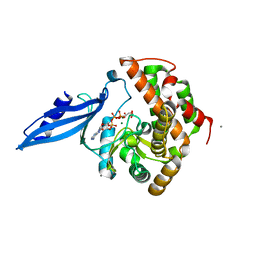 | | Crystal structure of Rv3168 with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kim, Y.-G, Kim, S, Nguyen, C.M.T, Kim, K.-J. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme
Proteins, 79, 2011
|
|
3ATS
 
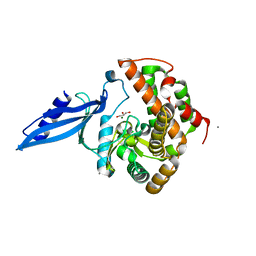 | | Crystal structure of Rv3168 | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y.-G, Kim, S, Nguyen, C.M.T, Kim, K.-J. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme
Proteins, 79, 2011
|
|
1NYJ
 
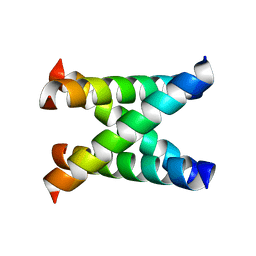 | |
6IM1
 
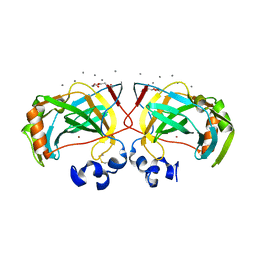 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), TETRAETHYLENE GLYCOL, ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
6IM0
 
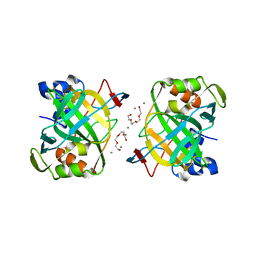 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | BICARBONATE ION, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
6IM3
 
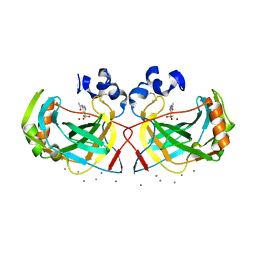 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
6JJL
 
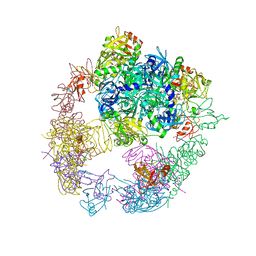 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-ARG-LYS-LEU, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy.
Commun Biol, 3, 2020
|
|
6JJK
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
6JJO
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | Periplasmic serine endoprotease DegP, TMB-CYRKL modulator | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.157 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
2JUA
 
 | | Assignment, structure, and dynamics of de novo designed protein S836 | | Descriptor: | de novo protein S836 | | Authors: | Go, A, Kim, S, Baum, J.S, Hecht, M.H. | | Deposit date: | 2007-08-16 | | Release date: | 2008-05-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of de novo proteins from a designed superfamily of 4-helix bundles.
Protein Sci., 17, 2008
|
|
3GWJ
 
 | | Crystal structure of Antheraea pernyi arylphorin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | Authors: | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
7DRN
 
 | | Structure of ATP-grasp ligase PsnB complexed with precursor peptide PsnA2 and AMPPNP | | Descriptor: | ATP-grasp domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PsnA214-38, ... | | Authors: | Song, I, Yu, J, Song, W, Kim, S. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
