2V7C
 
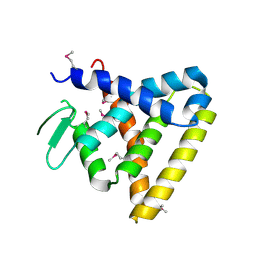 | | Crystal Structure of Rev-Erb beta | | Descriptor: | ORPHAN NUCLEAR RECEPTOR NR1D2 | | Authors: | Woo, E.-J, Jeong, D.G, Lim, M.-Y, Kim, S.J, Ryu, S.E. | | Deposit date: | 2007-07-29 | | Release date: | 2007-10-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into the Constitutive Repression Function of the Nuclear Receptor Rev-Erbbeta
J.Mol.Biol., 373, 2007
|
|
4JNB
 
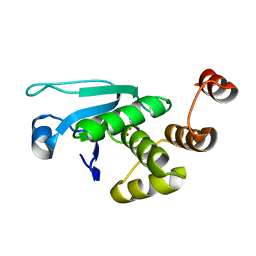 | | Crystal structure of the Catalytic Domain of Human DUSP12 | | Descriptor: | Dual specificity protein phosphatase 12, SULFATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JMJ
 
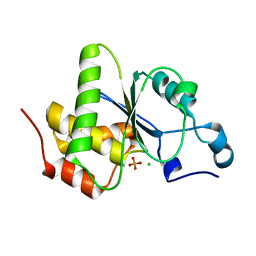 | | Structure of dusp11 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RNA/RNP complex-1-interacting phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JMK
 
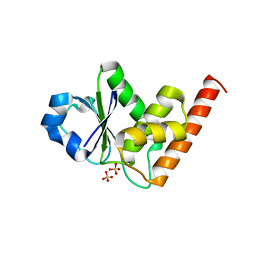 | | Structure of dusp8 | | Descriptor: | Dual specificity protein phosphatase 8, SULFATE ION | | Authors: | Jeong, D.G, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3HB1
 
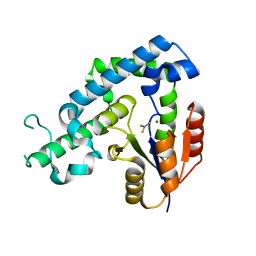 | | Crystal structure of ed-eya2 complexed with Alf3 | | Descriptor: | ALUMINUM FLUORIDE, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
3OIC
 
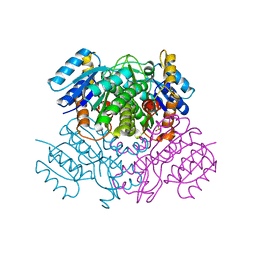 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], SULFATE ION | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OIG
 
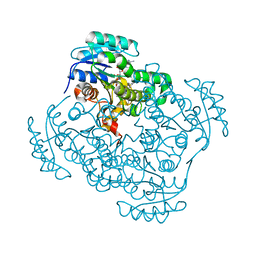 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and INH) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OID
 
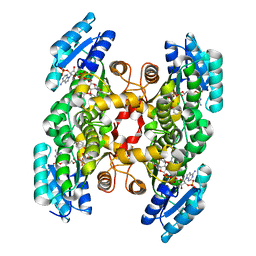 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (complex with NADP and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3OIF
 
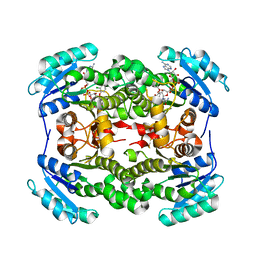 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
2ORY
 
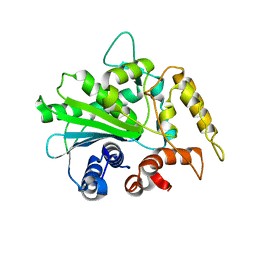 | | Crystal structure of M37 lipase | | Descriptor: | Lipase | | Authors: | Jung, S.K, Jeong, D.G, Kim, S.J. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum.
Proteins, 71, 2008
|
|
6IM5
 
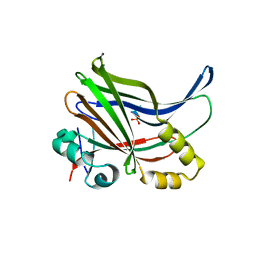 | | YAP-binding domain of human TEAD1 | | Descriptor: | PHOSPHATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Mo, Y, Lee, H.S, Kim, S.J, Ku, B. | | Deposit date: | 2018-10-22 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal Structure of the YAP-binding Domain of Human TEAD1
Bull.Korean Chem.Soc., 40, 2019
|
|
1XM2
 
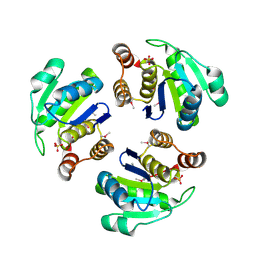 | | Crystal structure of Human PRL-1 | | Descriptor: | SULFATE ION, Tyrosine Phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Kim, J.H, Son, J.H, Ryu, S.E. | | Deposit date: | 2004-10-01 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms
J.Mol.Biol., 345, 2005
|
|
1ZZW
 
 | | Crystal Structure of catalytic domain of Human MAP Kinase Phosphatase 5 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 10, SULFATE ION | | Authors: | Jeong, D.G, Yoon, T.S, Kim, J.H, Shim, M.Y, Jeong, S.K, Son, J.H, Ryu, S.E, Kim, S.J. | | Deposit date: | 2005-06-14 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human MAP Kinase Phosphatase 5: Structural Insight into Constitutively Active Phosphatase.
J.Mol.Biol., 360, 2006
|
|
5CRV
 
 | | Crystal structure of the Bro domain of HD-PTP in a complex with the core region of STAM2 | | Descriptor: | GLYCEROL, Signal transducing adapter molecule 2, Tyrosine-protein phosphatase non-receptor type 23 | | Authors: | Lee, J, Ku, B, Kim, S.J. | | Deposit date: | 2015-07-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Study of the HD-PTP Bro1 Domain in a Complex with the Core Region of STAM2, a Subunit of ESCRT-0
Plos One, 11, 2016
|
|
5CRU
 
 | | Crystal structure of the Bro domain of HD-PTP | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 23 | | Authors: | Lee, J, Ku, B, Kim, S.J. | | Deposit date: | 2015-07-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Study of the HD-PTP Bro1 Domain in a Complex with the Core Region of STAM2, a Subunit of ESCRT-0
Plos One, 11, 2016
|
|
2NT2
 
 | | Crystal Structure of Slingshot phosphatase 2 | | Descriptor: | Protein phosphatase Slingshot homolog 2, SULFATE ION | | Authors: | Jung, S.K, Jeong, D.G, Yoon, T.S, Kim, J.H, Ryu, S.E, Kim, S.J. | | Deposit date: | 2006-11-06 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human slingshot phosphatase 2.
Proteins, 68, 2007
|
|
3EZZ
 
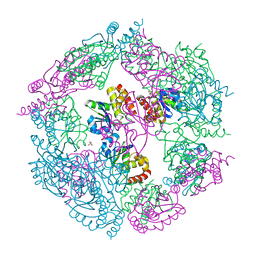 | | Crystal Structure of human MKP-2 | | Descriptor: | Dual specificity protein phosphatase 4, SULFATE ION | | Authors: | Jeong, D.G, Jung, S.K, Ryu, S.E, Kim, S.J. | | Deposit date: | 2008-10-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the catalytic domain of human MKP-2 reveals a 24-mer assembly.
Proteins, 76, 2009
|
|
4B04
 
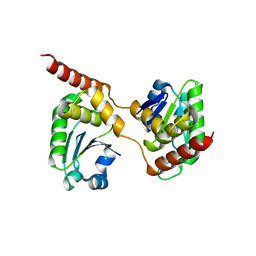 | | Crystal structure of the Catalytic Domain of Human DUSP26 (C152S) | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 26 | | Authors: | Won, E.-Y, Lee, D.Y, Park, S.G, Yokoyama, S, Kim, S.J, Chi, S.-W. | | Deposit date: | 2012-06-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | High-Resolution Crystal Structure of the Catalytic Domain of Human Dual-Specificity Phosphatase 26
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1UC7
 
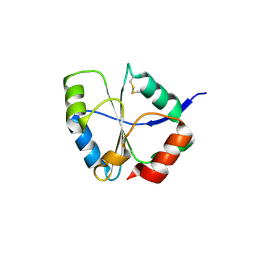 | | Crystal structure of DsbDgamma | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Kim, J.H, Kim, S.J, Jeong, D.G, Son, J.H, Ryu, S.E. | | Deposit date: | 2003-04-09 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of DsbDgamma reveals the mechanism of redox potential shift and substrate specificity(1)
FEBS LETT., 543, 2003
|
|
4HQR
 
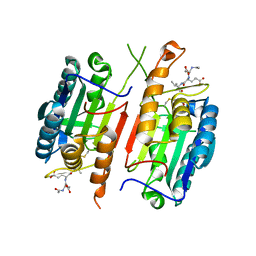 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Ac-Asp-Glu-Val-Asp-Aldehyde, Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-26 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HQ0
 
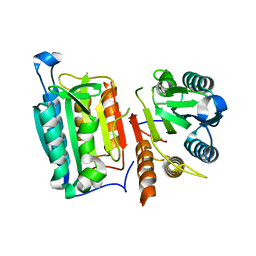 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-25 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3LJ8
 
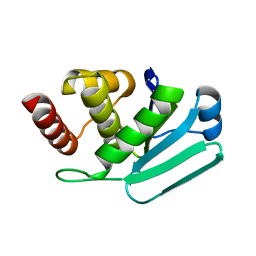 | | Crystal Structure of MKP-4 | | Descriptor: | Tyrosine-protein phosphatase | | Authors: | Jeong, D.G, Yoon, T.S, Jung, S.-K, Park, H.S, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring binding sites other than the catalytic core in the crystal structure of the catalytic domain of MKP-4
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3M7M
 
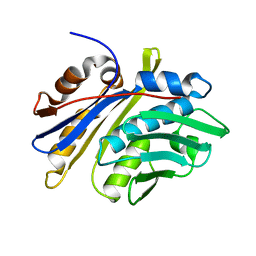 | | Crystal structure of monomeric hsp33 | | Descriptor: | 33 kDa chaperonin | | Authors: | Chi, S.W, Jeong, D.G, Woo, J.R, Park, B.C, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-03-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of monomeric hsp33
To be Published
|
|
3OBY
 
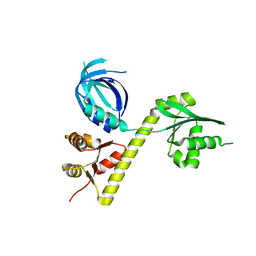 | | Crystal structure of Archaeoglobus fulgidus Pelota reveals inter-domain structural plasticity | | Descriptor: | Protein pelota homolog | | Authors: | Lee, H.H, Jang, J.Y, Yoon, H.-J, Kim, S.J, Suh, S.W. | | Deposit date: | 2010-08-09 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two archaeal Pelotas reveal inter-domain structural plasticity
Biochem.Biophys.Res.Commun., 399, 2010
|
|
