9IK1
 
 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | 登録日 | 2024-06-26 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
6L33
 
 | |
1HPB
 
 | |
2Q21
 
 | |
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | 分子名称: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | 著者 | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | 登録日 | 2014-09-04 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
1HP7
 
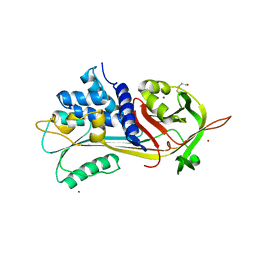 | | A 2.1 ANGSTROM STRUCTURE OF AN UNCLEAVED ALPHA-1-ANTITRYPSIN SHOWS VARIABILITY OF THE REACTIVE CENTER AND OTHER LOOPS | | 分子名称: | ALPHA-1-ANTITRYPSIN, BETA-MERCAPTOETHANOL, ZINC ION | | 著者 | Kim, S.-J, Woo, J.-R, Seo, E.J, Yu, M.-H, Ryu, S.-E. | | 登録日 | 2000-12-12 | | 公開日 | 2001-03-14 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A 2.1 A resolution structure of an uncleaved alpha(1)-antitrypsin shows variability of the reactive center and other loops.
J.Mol.Biol., 306, 2001
|
|
7Y28
 
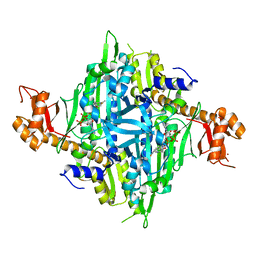 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | 分子名称: | 1-[6-(3-fluorophenyl)benzimidazol-1-yl]-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | 著者 | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | 登録日 | 2022-06-09 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y1H
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | 分子名称: | 1-(5-chloranyl-4-methyl-benzimidazol-1-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | 著者 | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | 登録日 | 2022-06-08 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y1W
 
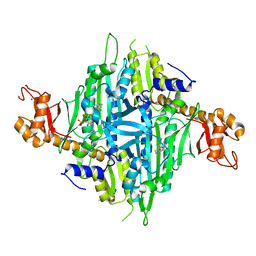 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | 分子名称: | (2R,3S)-2-[3-[4,5-bis(chloranyl)benzimidazol-1-yl]propyl]piperidin-3-ol, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | 著者 | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | 登録日 | 2022-06-09 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y3S
 
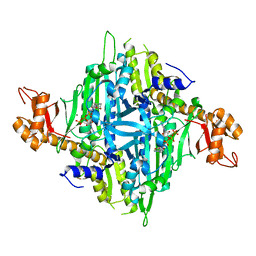 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | 分子名称: | 1-(6-bromanyl-7-methyl-imidazo[4,5-b]pyridin-3-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | 著者 | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | 登録日 | 2022-06-12 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
4WYS
 
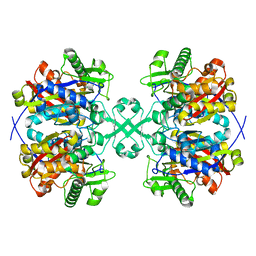 | | Crystal structure of thiolase from Escherichia coli | | 分子名称: | Acetyl-CoA acetyltransferase | | 著者 | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | 登録日 | 2014-11-18 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4WYR
 
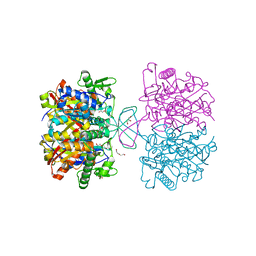 | | Crystal structure of thiolase mutation (V77Q,N153Y,A286K) from Clostridium acetobutylicum | | 分子名称: | Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | 著者 | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | 登録日 | 2014-11-18 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL4
 
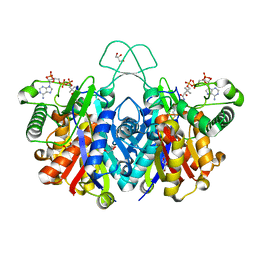 | | Crystal structure of thiolase from Clostridium acetobutylicum in complex with CoA | | 分子名称: | Acetyl-CoA acetyltransferase, COENZYME A, GLYCEROL | | 著者 | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | 登録日 | 2015-01-13 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL2
 
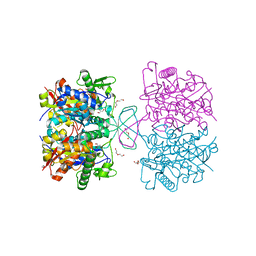 | | Crystal structure of oxidized form of thiolase from Clostridium acetobutylicum | | 分子名称: | ACETATE ION, Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | 登録日 | 2015-01-13 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL3
 
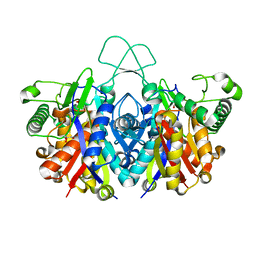 | | Crystal structure of reduced form of thiolase from Clostridium acetobutylicum | | 分子名称: | Acetyl-CoA acetyltransferase, GLYCEROL | | 著者 | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | 登録日 | 2015-01-13 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
7YA3
 
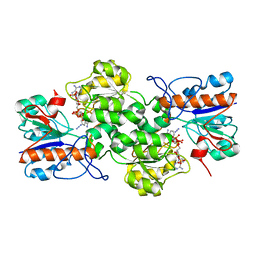 | |
7YA4
 
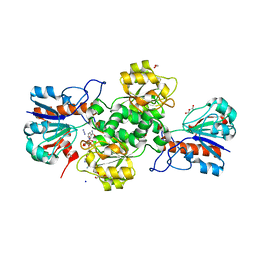 | |
8I70
 
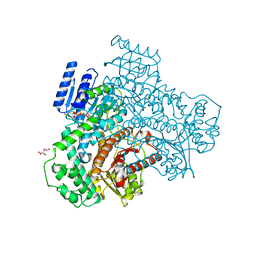 | |
4XGS
 
 | |
8I6Z
 
 | |
132D
 
 | |
7VG4
 
 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 strain | | 分子名称: | Methenyltetrahydrofolate cyclohydrolase | | 著者 | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | 登録日 | 2021-09-14 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
7VG5
 
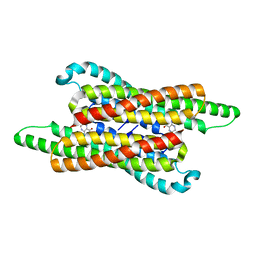 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 with tetrahydrofolate | | 分子名称: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Methenyltetrahydrofolate cyclohydrolase | | 著者 | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | 登録日 | 2021-09-14 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
4JDY
 
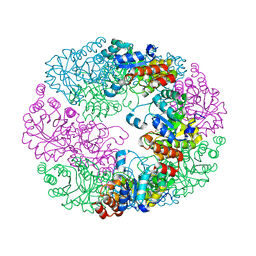 | | Crystal structure of Rv2606c | | 分子名称: | GLYCEROL, Pyridoxal biosynthesis lyase PdxS | | 著者 | Kim, S, Kim, K.-J. | | 登録日 | 2013-02-25 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis Rv2606c: a pyridoxal biosynthesis lyase.
Biochem.Biophys.Res.Commun., 435, 2013
|
|
4KDL
 
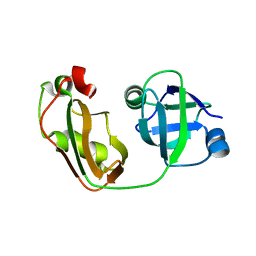 | | Crystal structure of p97/VCP N in complex with OTU1 UBXL | | 分子名称: | Transitional endoplasmic reticulum ATPase, Ubiquitin thioesterase OTU1 | | 著者 | Kim, S.J, Kim, E.E. | | 登録日 | 2013-04-25 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural Basis for Ovarian Tumor Domain-containing Protein 1 (OTU1) Binding to p97/Valosin-containing Protein (VCP).
J.Biol.Chem., 289, 2014
|
|
