1VLS
 
 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR | | Descriptor: | ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
1VLT
 
 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR WITH ASPARTATE | | Descriptor: | ASPARTATE RECEPTOR, ASPARTIC ACID | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
5Y9D
 
 | | Crystal structure of acyl-coA oxidase1 from Yarrowia lipolytica | | Descriptor: | Acyl-coenzyme A oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2017-08-24 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the substrate specificity of acyl-CoA oxidase1 from Yarrowia lipolytica for short-chain dicarboxylyl-CoAs.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
2LIG
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR, ASPARTIC ACID, ... | | Authors: | Kim, S.-H, Yeh, J.I, Prive, G.G, Milburn, M, Scott, W, Koshland Junior, D.E. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
2LAO
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN WITH AND WITHOUT A LIGAND | | Descriptor: | LYSINE, ARGININE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H, Kang, C.-H. | | Deposit date: | 1993-02-25 | | Release date: | 1994-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand.
J.Biol.Chem., 268, 1993
|
|
9IK1
 
 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | Descriptor: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | Authors: | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2014-09-04 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
7Y1W
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | (2R,3S)-2-[3-[4,5-bis(chloranyl)benzimidazol-1-yl]propyl]piperidin-3-ol, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y1H
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(5-chloranyl-4-methyl-benzimidazol-1-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y28
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-[6-(3-fluorophenyl)benzimidazol-1-yl]-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y3S
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(6-bromanyl-7-methyl-imidazo[4,5-b]pyridin-3-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-12 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
4WYS
 
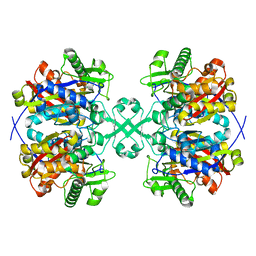 | | Crystal structure of thiolase from Escherichia coli | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4WYR
 
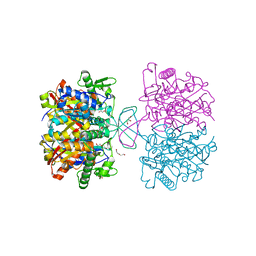 | | Crystal structure of thiolase mutation (V77Q,N153Y,A286K) from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
9JO4
 
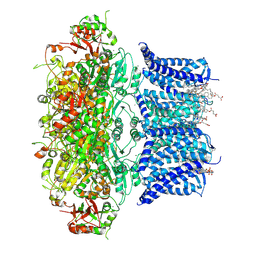 | | Cryo-EM structure of human BKca channel-compound 51b complex | | Descriptor: | 5-(5-morpholin-4-ylpentylamino)-2-[2,3,5,6-tetrakis(fluoranyl)-4-(trifluoromethyl)phenoxy]phenol, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kim, S, Park, S, Lee, N.Y, Lee, E.Y, Lee, N, Roh, E.C, Kim, Y.G, Kim, H.J, Jin, M.S, Park, C.S, Kim, Y.C. | | Deposit date: | 2024-09-24 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery of Diphenyl Ether Derivatives as Novel BK Ca Channel Activators: Structure-Activity Relationship, Cryo-EM Complex Structures, and In Vivo Animal Studies.
J.Med.Chem., 68, 2025
|
|
9JO3
 
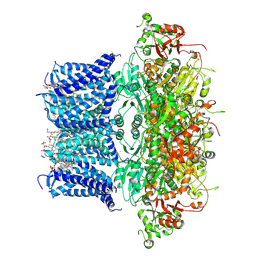 | | Cryo-EM structure of human BKca channel-compound 10b complex | | Descriptor: | 5-azanyl-2-[2,3,5,6-tetrakis(fluoranyl)-4-(trifluoromethyl)phenoxy]phenol, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kim, S, Park, S, Lee, N.Y, Lee, E.Y, Lee, N, Roh, E.C, Kim, Y.G, Kim, H.J, Jin, M.S, Park, C.S, Kim, Y.C. | | Deposit date: | 2024-09-24 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery of Diphenyl Ether Derivatives as Novel BK Ca Channel Activators: Structure-Activity Relationship, Cryo-EM Complex Structures, and In Vivo Animal Studies.
J.Med.Chem., 68, 2025
|
|
4XL2
 
 | | Crystal structure of oxidized form of thiolase from Clostridium acetobutylicum | | Descriptor: | ACETATE ION, Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL3
 
 | | Crystal structure of reduced form of thiolase from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL4
 
 | | Crystal structure of thiolase from Clostridium acetobutylicum in complex with CoA | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
7YA3
 
 | |
7R22
 
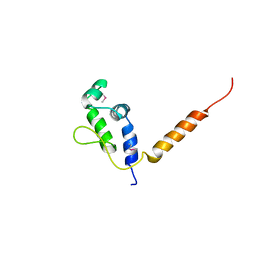 | |
7YA4
 
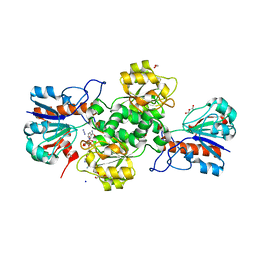 | |
9LSK
 
 | | Cryo-EM structure of the Klebsiella pneumoniae CitS (citrate-bound occluded state) | | Descriptor: | CITRIC ACID, Citrate/sodium symporter, PALMITIC ACID | | Authors: | Kim, S, Kim, J.W, Park, J.G, Lee, S.S, Choi, S.H, Lee, J.-O, Jin, M.S. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Disulfide-stabilized diabodies enable near-atomic cryo-EM imaging of small proteins: A case study of the bacterial Na + /citrate symporter CitS.
Structure, 2025
|
|
9LSJ
 
 | | Cryo-EM structure of the G15C-R66C and T83C-T83C diabody complex (CitS-diabody #7-TLR3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Kim, S, Kim, J.W, Park, J.G, Lee, S.S, Choi, S.H, Lee, J.-O, Jin, M.S. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Disulfide-stabilized diabodies enable near-atomic cryo-EM imaging of small proteins: A case study of the bacterial Na + /citrate symporter CitS.
Structure, 2025
|
|
9LSI
 
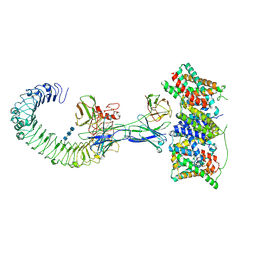 | | Cryo-EM structure of the S82C-S82C diabody complex (CitS-diabody #2-TLR3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, S, Kim, J.W, Park, J.G, Lee, S.S, Choi, S.H, Lee, J.-O, Jin, M.S. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Disulfide-stabilized diabodies enable near-atomic cryo-EM imaging of small proteins: A case study of the bacterial Na + /citrate symporter CitS.
Structure, 2025
|
|
9LSH
 
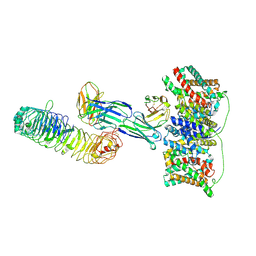 | | Cryo-EM structure of the wild-type diabody complex (CitS-diabody #1-TLR3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, S, Kim, J.W, Park, J.G, Lee, S.S, Choi, S.H, Lee, J.-O, Jin, M.S. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Disulfide-stabilized diabodies enable near-atomic cryo-EM imaging of small proteins: A case study of the bacterial Na + /citrate symporter CitS.
Structure, 2025
|
|
