1MSH
 
 | |
3BVE
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
1NCO
 
 | | STRUCTURE OF THE ANTITUMOR PROTEIN-CHROMOPHORE COMPLEX NEOCARZINOSTATIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, HOLO-NEOCARZINOSTATIN, NEOCARZINOSTATIN-CHROMOPHORE | | Authors: | Kim, K.-H, Kwon, B.-M, Myers, A.G, Rees, D.C. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of neocarzinostatin, an antitumor protein-chromophore complex.
Science, 262, 1993
|
|
3NAI
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | 5-FLUOROURACIL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
3NAH
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | RNA dependent RNA polymerase, SULFATE ION | | Authors: | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
3SFU
 
 | | crystal structure of murine norovirus RNA dependent RNA polymerase in complex with ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Alam, I. | | Deposit date: | 2011-06-14 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin.
Virology, 426, 2012
|
|
3SFG
 
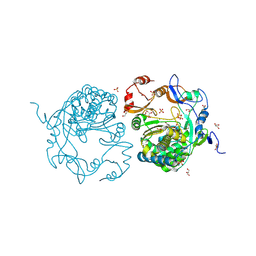 | |
3QC8
 
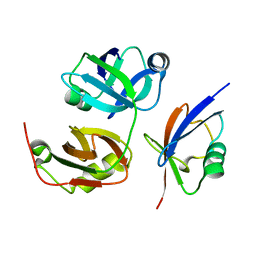 | | Crystal Structure of FAF1 UBX Domain In Complex with p97/VCP N Domain Reveals The Conserved FcisP Touch-Turn Motif of UBX Domain Suffering Conformational Change | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Kim, K.H, Kang, W, Suh, S.W, Yang, J.K. | | Deposit date: | 2011-01-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain
Proteins, 79, 2011
|
|
3QID
 
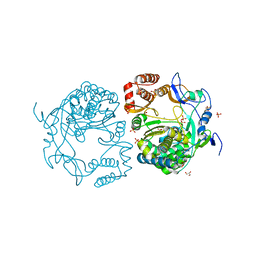 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
2Q37
 
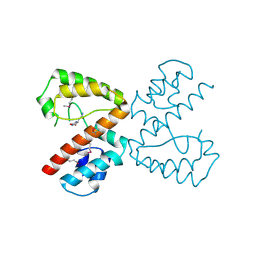 | |
3P1L
 
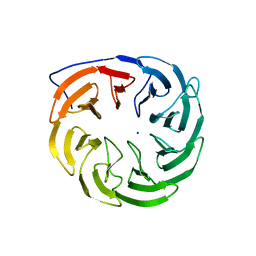 | | Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex, native crystals. | | Descriptor: | Lipoprotein yfgL, SODIUM ION | | Authors: | Kim, K.H, Paetzel, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex, native crystals
J.Mol.Biol., 406, 2011
|
|
4QB9
 
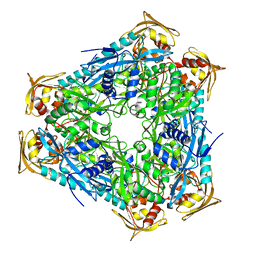 | | Crystal structure of Mycobacterium smegmatis Eis in complex with paromomycin | | Descriptor: | Enhanced intracellular survival protein, PAROMOMYCIN, SULFATE ION | | Authors: | Kim, K.H, Ahn, D.R, Yoon, H.J, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structure of Mycobacterium smegmatis Eis in complex with paromomycin.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3S2P
 
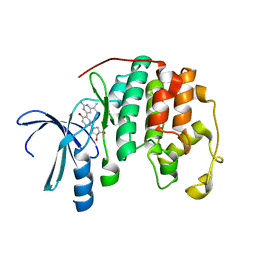 | | Crystal structure of CDK2 with a 2-aminopyrimidine compound | | Descriptor: | (3S,4S)-1-{3-[2-amino-6-(propan-2-yl)pyrimidin-4-yl]-4-hydroxyphenyl}pyrrolidine-3,4-diol, Cyclin-dependent kinase 2 | | Authors: | Kim, K.-H, Lee, J, Jeong, S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of 2-aminopyrimidines as CDK1 and CDK2 inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SNS
 
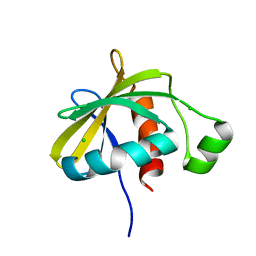 | | Crystal structure of the C-terminal domain of Escherichia coli lipoprotein BamC | | Descriptor: | CHLORIDE ION, Lipoprotein 34 | | Authors: | Kim, K.H, Aulakh, S, Tan, W, Paetzel, M. | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic analysis of the C-terminal domain of the Escherichia coli lipoprotein BamC.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5Z9G
 
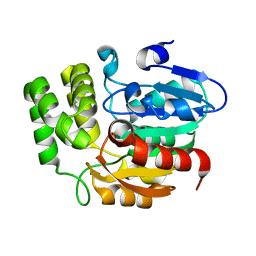 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5Z9H
 
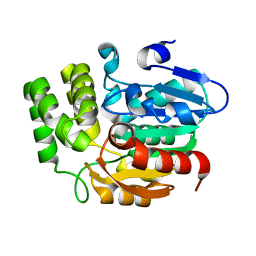 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
7CMY
 
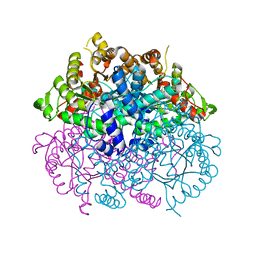 | | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate | | Descriptor: | GLYOXYLIC ACID, Isocitrate lyase, MAGNESIUM ION, ... | | Authors: | Kim, K, Ki, D, Lee, S.H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate
To Be Published
|
|
7CMX
 
 | |
5Y86
 
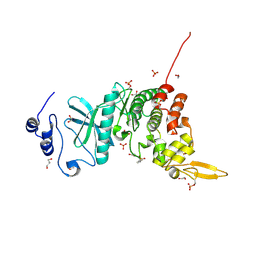 | | Crystal structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 3, ... | | Authors: | Kim, K.L, Cha, J.S, Cho, Y.S, Kim, H.Y, Chang, N.P, Cho, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
J. Mol. Biol., 430, 2018
|
|
5Y3D
 
 | |
5XDA
 
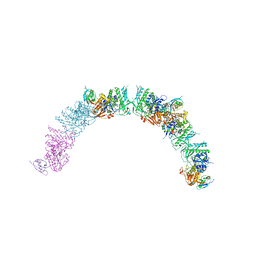 | |
5ZU1
 
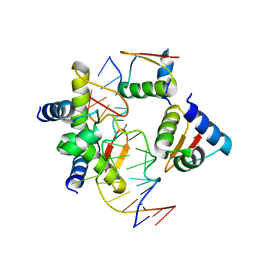 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*AP*GP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*TP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-05 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
6AE3
 
 | | Crystal structure of GSK3beta complexed with Morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, GLYCEROL, Glycogen synthase kinase-3 beta | | Authors: | Kim, K.L, Cha, J.S, Kim, J.S, Ahn, J.S, Ha, N.C, Cho, H.S. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of GSK3 beta in complex with the flavonoid, morin
Biochem. Biophys. Res. Commun., 504, 2018
|
|
5ZUO
 
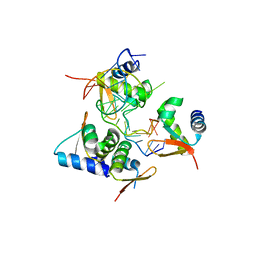 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5ZUP
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*AP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
