6KZ7
 
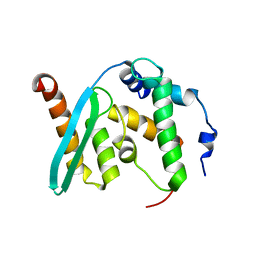 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
6O9Y
 
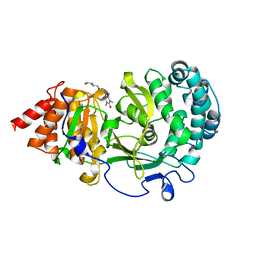 | | Structure of human PARG complexed with JA2-8 | | Descriptor: | 7-[(2S)-2-hydroxy-3-(morpholin-4-yl)propyl]-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6OA3
 
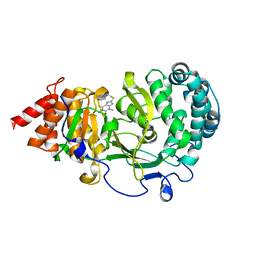 | | Structure of human PARG complexed with JA2131 | | Descriptor: | (8S)-1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,7,8,9-hexahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7LBE
 
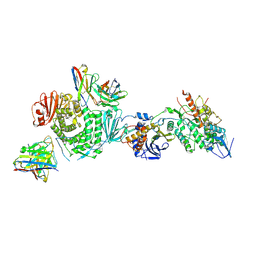 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBG
 
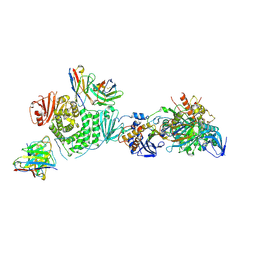 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
6OA0
 
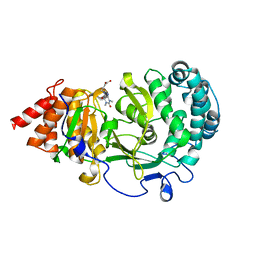 | | Structure of human PARG complexed with JA2-9 | | Descriptor: | 4-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)butanoic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7LBF
 
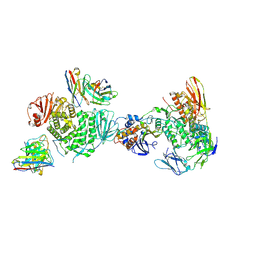 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
2JYP
 
 | |
2A4V
 
 | | Crystal Structure of a truncated mutant of yeast nuclear thiol peroxidase | | Descriptor: | Peroxiredoxin DOT5 | | Authors: | Choi, J, Choi, S, Chon, J.-K, Choi, J, Cha, M.-K, Kim, I.-H, Shin, W. | | Deposit date: | 2005-06-29 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C107S/C112S mutant of yeast nuclear 2-Cys peroxiredoxin
Proteins, 61, 2005
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
1FNX
 
 | | SOLUTION STRUCTURE OF THE HUC RBD1-RBD2 COMPLEXED WITH THE AU-RICH ELEMENT | | Descriptor: | AU-RICH RNA ELEMENT, HU ANTIGEN C | | Authors: | Inoue, M, Hirao, M, Kasashima, K, Kim, I.-S, Kawai, G, Kigawa, T, Sakamoto, H, Muto, Y, Yokoyama, S. | | Deposit date: | 2000-08-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse HuC RNA-binding domains complexed with an AU-Rich element reveals determinants of neuronal differentiation
To be Published, 2000
|
|
1P5M
 
 | |
1P5O
 
 | |
1P5P
 
 | |
1P5N
 
 | |
5XN4
 
 | | Anti-CRISPR protein AcrIIA4 | | Descriptor: | Anti-CRISPR AcrIIA4 | | Authors: | Suh, J.-Y, Kim, I. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor.
Sci Rep, 8, 2018
|
|
8V9A
 
 | | GII.NA1 Loreto 1257 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1 | | Authors: | Kher, G, Kim, I, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
5Z6V
 
 | |
5Y4J
 
 | |
5Y4I
 
 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8R
 
 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
